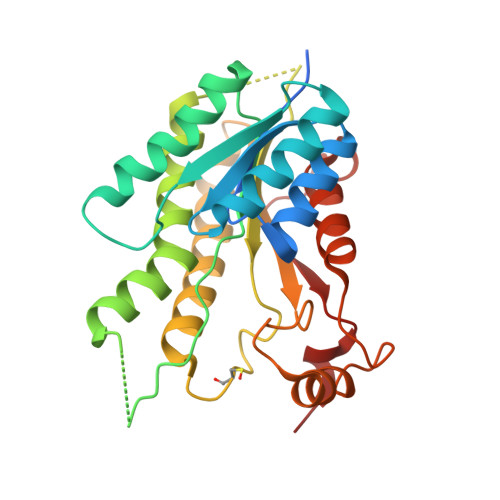
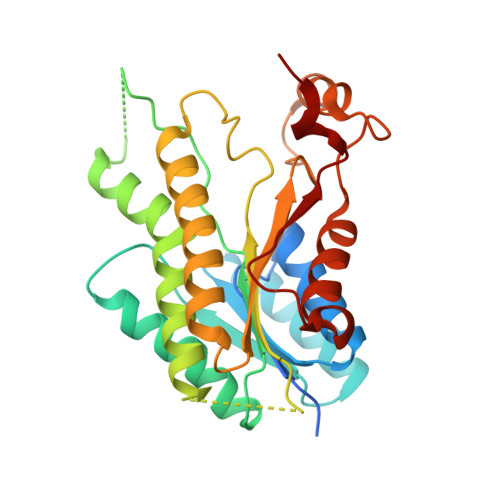
Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
Khalaf, A.I., Huggan, J.K., Suckling, C.J., Gibson, C.L., Stewart, K., Giordani, F., Barrett, M.P., Wong, P.E., Barrack, K.L., Hunter, W.N.(2014) J Med Chem 57: 6479
- PubMed: 25007262
- DOI: https://doi.org/10.1021/jm500483b
- Primary Citation of Related Structures:
4CL8, 4CLD, 4CLE, 4CLH, 4CLO, 4CLR, 4CLX, 4CM1, 4CM3, 4CM4, 4CM5, 4CM6, 4CM7, 4CM8, 4CM9, 4CMA, 4CMB, 4CMC, 4CME, 4CMG, 4CMI, 4CMJ, 4CMK - PubMed Abstract:
The treatment of Human African trypanosomiasis remains a major unmet health need in sub-Saharan Africa. Approaches involving new molecular targets are important; pteridine reductase 1 (PTR1), an enzyme that reduces dihydrobiopterin in Trypanosoma spp., has been identified as a candidate target, and it has been shown previously that substituted pyrrolo[2,3-d]pyrimidines are inhibitors of PTR1 from Trypanosoma brucei (J. Med. Chem. 2010, 53, 221-229). In this study, 61 new pyrrolo[2,3-d]pyrimidines have been prepared, designed with input from new crystal structures of 23 of these compounds complexed with PTR1, and evaluated in screens for enzyme inhibitory activity against PTR1 and in vitro antitrypanosomal activity. Eight compounds were sufficiently active in both screens to take forward to in vivo evaluation. Thus, although evidence for trypanocidal activity in a stage I disease model in mice was obtained, the compounds were too toxic to mice for further development.
- WestCHEM Department of Pure and Applied Chemistry, University of Strathclyde , 295 Cathedral Street, Glasgow G1 1XL, United Kingdom.
Organizational Affiliation: