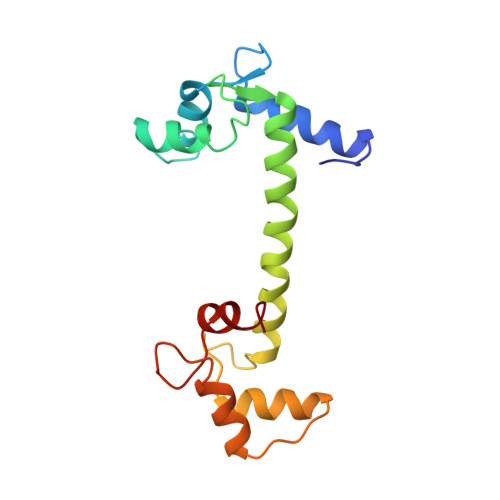
Structure of a recombinant calmodulin from Drosophila melanogaster refined at 2.2-A resolution.
Taylor, D.A., Sack, J.S., Maune, J.F., Beckingham, K., Quiocho, F.A.(1991) J Biological Chem 266: 21375-21380
- PubMed: 1939171
- DOI: https://doi.org/10.2210/pdb4cln/pdb
- Primary Citation of Related Structures:
4CLN - PubMed Abstract:
The crystal structure of calmodulin (Mr 16,700, 148 residues) from Drosophila melanogaster as expressed in a bacterial system has been determined and refined at 2.2-A resolution. Starting with the structure of mammalian calmodulin, we produced an extensively refitted and refined model with a conventional crystallographic R value of 0.197 for the 5,239 reflections (F greater than or equal to 2 sigma (F)) within the 10.0-2.2-A resolution range. The model includes 1,164 protein atoms, 4 calcium ions, and 78 water molecules and has root mean square deviations from standard values of 0.018 A for bond lengths and 0.043 A for angle distances. The overall structure is similar to mammalian calmodulin, with a seven-turn central helix connecting the two calcium-binding domains. The "dumb-bell" shaped molecule contains seven alpha-helices and four "EF hand" calcium-binding sites. Although the amino acid sequences of mammalian and Drosophila calmodulins differ by only three conservative amino acid changes, the refined model reveals a number of significant differences between the two structures. Superimposition of the structures yields a root mean square deviation of 1.22 A for the 1,120 equivalent atoms. The calcium-binding domains have a root mean square deviation of 0.85 A for the 353 equivalent atoms. There are also differences in the amino terminus, the bend of the central alpha-helix, and the orientations of some of the side chains.
- Howard Hughes Medical Institute, Houston, Texas.
Organizational Affiliation: