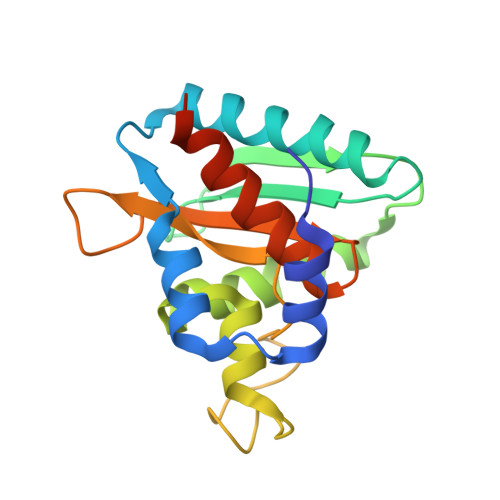
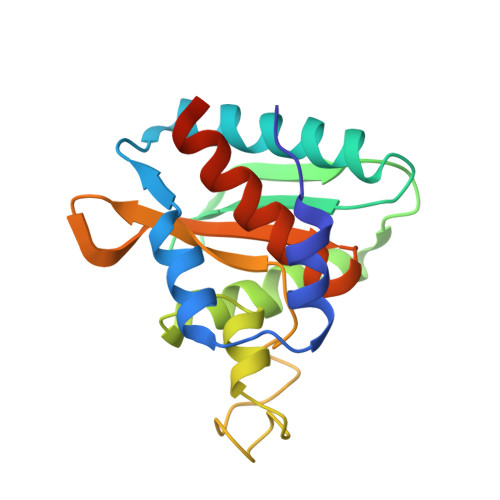
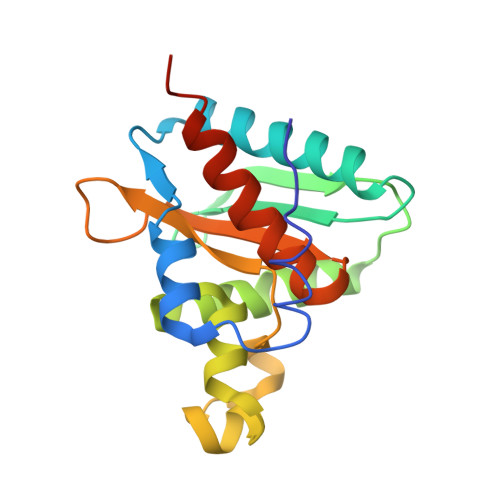
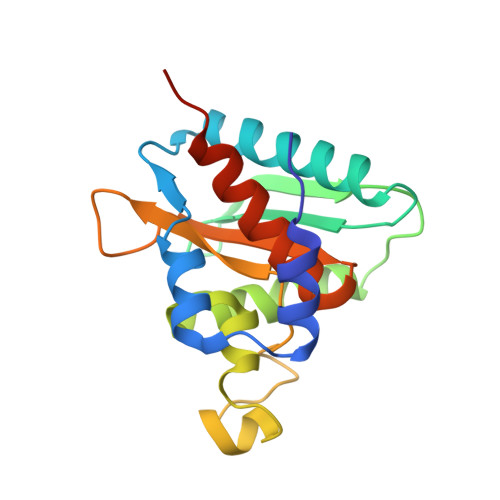
Structural and Functional Characterization of Ybr137Wp Implicate its Involvement in the Targeting of Tail-Anchored Proteins to Membranes.
Yeh, Y., Lin, T., Li, Y., Tung, J., Lin, C., Hsiao, C.(2014) Mol Cell Biol 34: 4500
- PubMed: 25288638
- DOI: https://doi.org/10.1128/MCB.00697-14
- Primary Citation of Related Structures:
4CLC - PubMed Abstract:
Nearly 5% of membrane proteins are guided to nuclear, endoplasmic reticulum (ER), mitochondrial, Golgi, or peroxisome membranes by their C-terminal transmembrane domain and are classified as tail-anchored (TA) membrane proteins. In Saccharomyces cerevisiae, the guided entry of TA protein (GET) pathway has been shown to function in the delivery of TA proteins to the ER. The sorting complex for this pathway is comprised of Sgt2, Get4, and Get5 and facilitates the loading of nascent tail-anchored proteins onto the Get3 ATPase. Multiple pulldown assays also indicated that Ybr137wp associates with this complex in vivo. Here, we report a 2.8-Å-resolution crystal structure for Ybr137wp from Saccharomyces cerevisiae. The protein is a decamer in the crystal and also in solution, as observed by size exclusion chromatography and analytical ultracentrifugation. In addition, isothermal titration calorimetry indicated that the C-terminal acidic motif of Ybr137wp interacts with the tetratricopeptide repeat (TPR) domain of Sgt2. Moreover, an in vivo study demonstrated that Ybr137wp is induced in yeast exiting the log phase and ameliorates the defect of TA protein delivery and cell viability derived by the impaired GET system under starvation conditions. Therefore, this study suggests a possible role for Ybr137wp related to targeting of tail-anchored proteins.
- Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan.
Organizational Affiliation: