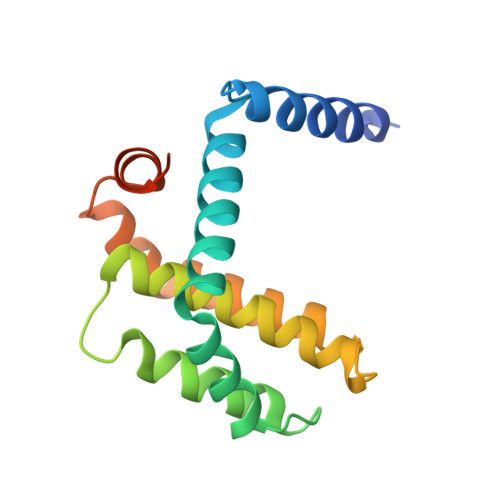
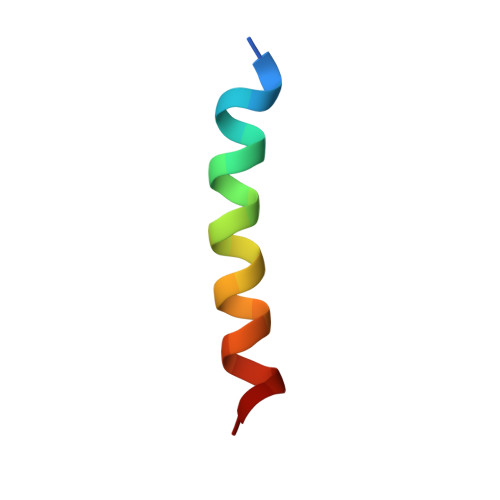
The Functional Differences of Pro-Survival and Pro-Apoptotic B Cell Lymphoma 2 (Bcl-2) Proteins Depend on Structural Differences in Their Bcl-2 Homology 3 (Bh3) Domains
Lee, E.F., Dewson, G., Evangelista, M., Pettikiriarachchi, A., Zhu, H., Colman, P.M., Fairlie, W.D.(2014) J Biological Chem 289: 36001
- PubMed: 25371206
- DOI: https://doi.org/10.1074/jbc.M114.610758
- Primary Citation of Related Structures:
4CIM, 4CIN - PubMed Abstract:
Bcl-2 homology 3 (BH3) domains are short sequence motifs that mediate nearly all protein-protein interactions between B cell lymphoma 2 (Bcl-2) family proteins in the intrinsic apoptotic cell death pathway. These sequences are found on both pro-survival and pro-apoptotic members, although their primary function is believed to be associated with induction of cell death. Here, we identify critical features of the BH3 domains of pro-survival proteins that distinguish them functionally from their pro-apoptotic counterparts. Biochemical and x-ray crystallographic studies demonstrate that these differences reduce the capacity of most pro-survival proteins to form high affinity "BH3-in-groove" complexes that are critical for cell death induction. Switching these residues for the corresponding residues in Bcl-2 homologous antagonist/killer (Bak) increases the binding affinity of isolated BH3 domains for pro-survival proteins; however, their exchange in the context of the parental protein causes rapid proteasomal degradation due to protein destabilization. This is supported by further x-ray crystallographic studies that capture elements of this destabilization in one pro-survival protein, Bcl-w. In pro-apoptotic Bak, we demonstrate that the corresponding distinguishing residues are important for its cell-killing capacity and antagonism by pro-survival proteins.
- From the Walter and Eliza Hall Institute of Medical Research, 1G Royal Pde, Parkville, Victoria 3052, Australia and the Department of Medical Biology, University of Melbourne, Parkville, Victoria 3010, Australia.
Organizational Affiliation: