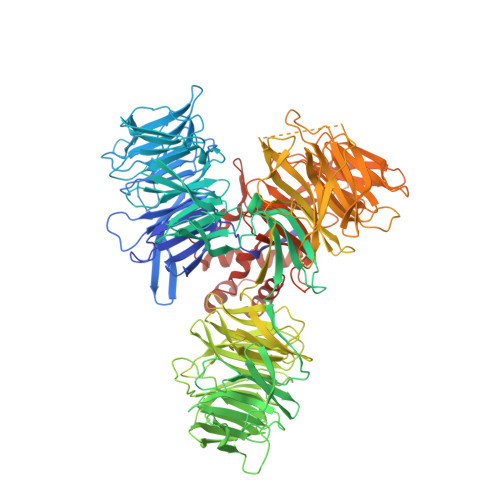
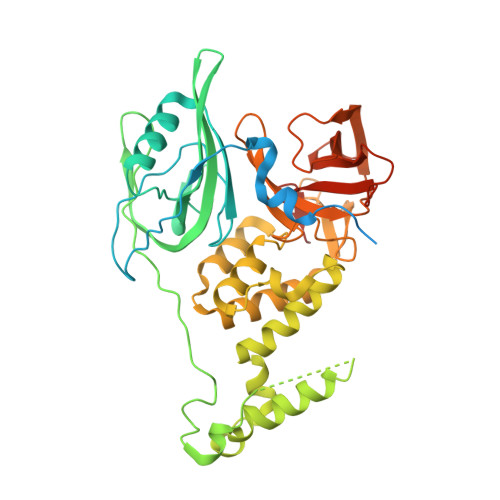
Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Fischer, E.S., Bohm, K., Lydeard, J.R., Yang, H., Stadler, M.B., Cavadini, S., Nagel, J., Serluca, F., Acker, V., Lingaraju, G.M., Tichkule, R.B., Schebesta, M., Forrester, W.C., Schirle, M., Hassiepen, U., Ottl, J., Hild, M., Beckwith, R.E.J., Harper, J.W., Jenkins, J.L., Thoma, N.H.(2014) Nature 512: 49
- PubMed: 25043012
- DOI: https://doi.org/10.1038/nature13527
- Primary Citation of Related Structures:
4CI1, 4CI2, 4CI3 - PubMed Abstract:
In the 1950s, the drug thalidomide, administered as a sedative to pregnant women, led to the birth of thousands of children with multiple defects. Despite the teratogenicity of thalidomide and its derivatives lenalidomide and pomalidomide, these immunomodulatory drugs (IMiDs) recently emerged as effective treatments for multiple myeloma and 5q-deletion-associated dysplasia. IMiDs target the E3 ubiquitin ligase CUL4-RBX1-DDB1-CRBN (known as CRL4(CRBN)) and promote the ubiquitination of the IKAROS family transcription factors IKZF1 and IKZF3 by CRL4(CRBN). Here we present crystal structures of the DDB1-CRBN complex bound to thalidomide, lenalidomide and pomalidomide. The structure establishes that CRBN is a substrate receptor within CRL4(CRBN) and enantioselectively binds IMiDs. Using an unbiased screen, we identified the homeobox transcription factor MEIS2 as an endogenous substrate of CRL4(CRBN). Our studies suggest that IMiDs block endogenous substrates (MEIS2) from binding to CRL4(CRBN) while the ligase complex is recruiting IKZF1 or IKZF3 for degradation. This dual activity implies that small molecules can modulate an E3 ubiquitin ligase and thereby upregulate or downregulate the ubiquitination of proteins.
- 1] Friedrich Miescher Institute for Biomedical Research, Maulbeerstrasse 66, CH-4058 Basel, Switzerland [2] University of Basel, Petersplatz 10, CH-4003 Basel, Switzerland.
Organizational Affiliation: