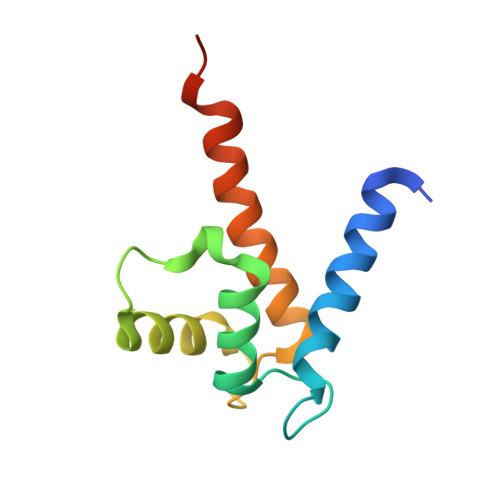
The C-Terminal Random Coil Region Tunes the Ca2+-Binding Affinity of S100A4 Through Conformational Activation.
Duelli, A., Kiss, B., Lundholm, I., Bodor, A., Radnai, L., Petoukhov, M., Svergun, D., Nyitray, L., Katona, G.(2014) PLoS One 9: 97654
- PubMed: 24830809
- DOI: https://doi.org/10.1371/journal.pone.0097654
- Primary Citation of Related Structures:
4CFQ, 4CFR - PubMed Abstract:
S100A4 interacts with many binding partners upon Ca2+ activation and is strongly associated with increased metastasis formation. In order to understand the role of the C-terminal random coil for the protein function we examined how small angle X-ray scattering of the wild-type S100A4 and its C-terminal deletion mutant (residues 1-88, Δ13) changes upon Ca2+ binding. We found that the scattering intensity of wild-type S100A4 changes substantially in the 0.15-0.25 Å-1 q-range whereas a similar change is not visible in the C-terminus deleted mutant. Ensemble optimization SAXS modeling indicates that the entire C-terminus is extended when Ca2+ is bound. Pulsed field gradient NMR measurements provide further support as the hydrodynamic radius in the wild-type protein increases upon Ca2+ binding while the radius of Δ13 mutant does not change. Molecular dynamics simulations provide a rational explanation of the structural transition: the positively charged C-terminal residues associate with the negatively charged residues of the Ca2+-free EF-hands and these interactions loosen up considerably upon Ca2+-binding. As a consequence the Δ13 mutant has increased Ca2+ affinity and is constantly loaded at Ca2+ concentration ranges typically present in cells. The activation of the entire C-terminal random coil may play a role in mediating interaction with selected partner proteins of S100A4.
- Department of Chemistry and Molecular Biology, University of Gothenburg, Gothenburg, Sweden.
Organizational Affiliation: