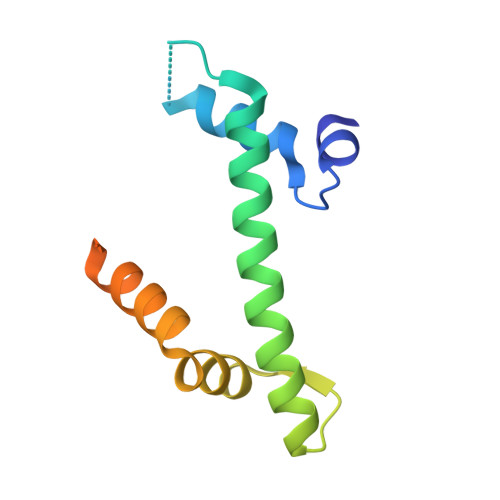
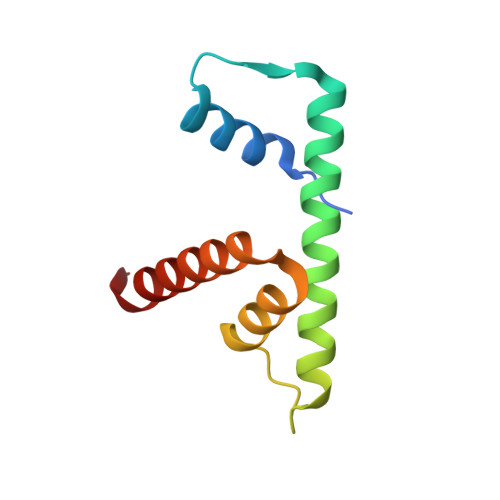
Anp32E is a Histone Chaperone that Removes H2A.Z from Chromatin
Obri, A., Ouararhni, K., Papin, C., Diebold, M.-L., Padmanabhan, K., Marek, M., Stoll, I., Roy, L., Reilly, P.T., Wak, T.W., Dimitrov, S., Romier, C., Hamiche, A.(2014) Nature 648: 505
- PubMed: 24463511
- DOI: https://doi.org/10.1038/nature12922
- Primary Citation of Related Structures:
4CAY - PubMed Abstract:
H2A.Z is an essential histone variant implicated in the regulation of key nuclear events. However, the metazoan chaperones responsible for H2A.Z deposition and its removal from chromatin remain unknown. Here we report the identification and characterization of the human protein ANP32E as a specific H2A.Z chaperone. We show that ANP32E is a member of the presumed H2A.Z histone-exchange complex p400/TIP60. ANP32E interacts with a short region of the docking domain of H2A.Z through a new motif termed H2A.Z interacting domain (ZID). The 1.48 Å resolution crystal structure of the complex formed between the ANP32E-ZID and the H2A.Z/H2B dimer and biochemical data support an underlying molecular mechanism for H2A.Z/H2B eviction from the nucleosome and its stabilization by ANP32E through a specific extension of the H2A.Z carboxy-terminal α-helix. Finally, analysis of H2A.Z localization in ANP32E(-/-) cells by chromatin immunoprecipitation followed by sequencing shows genome-wide enrichment, redistribution and accumulation of H2A.Z at specific chromatin control regions, in particular at enhancers and insulators.
- 1] Département de Génomique Fonctionnelle et Cancer, Institut de Génétique et Biologie Moléculaire et Cellulaire (IGBMC), Université de Strasbourg, CNRS, INSERM, 1 rue Laurent Fries, B.P. 10142, 67404 Illkirch Cedex, France [2].
Organizational Affiliation: