Mechanism of Farnesylated Caax Protein Processing by the Intramembrane Protease Rce1
Manolaridis, I., Kulkarni, K., Dodd, R.B., Ogasawara, S., Zhang, Z., Bineva, G., O'Reilly, N., Hanrahan, S.J., Thompson, A.J., Cronin, N., Iwata, S., Barford, D.(2013) Nature 504: 301
- PubMed: 24291792
- DOI: https://doi.org/10.1038/nature12754
- Primary Citation of Related Structures:
4CAD - PubMed Abstract:
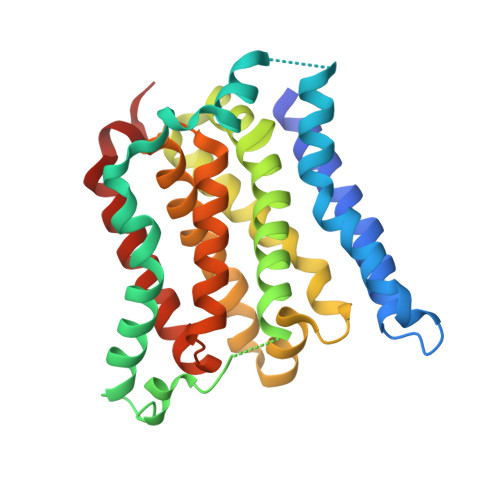
CAAX proteins have essential roles in multiple signalling pathways, controlling processes such as proliferation, differentiation and carcinogenesis. The ∼120 mammalian CAAX proteins function at cellular membranes and include the Ras superfamily of small GTPases, nuclear lamins, the γ-subunit of heterotrimeric GTPases, and several protein kinases and phosphatases. The proper localization of CAAX proteins to cell membranes is orchestrated by a series of post-translational modifications of the carboxy-terminal CAAX motifs (where C is cysteine, A is an aliphatic amino acid and X is any amino acid). These reactions involve prenylation of the cysteine residue, cleavage at the AAX tripeptide and methylation of the carboxyl-prenylated cysteine residue. The major CAAX protease activity is mediated by Rce1 (Ras and a-factor converting enzyme 1), an intramembrane protease (IMP) of the endoplasmic reticulum. Information on the architecture and proteolytic mechanism of Rce1 has been lacking. Here we report the crystal structure of a Methanococcus maripaludis homologue of Rce1, whose endopeptidase specificity for farnesylated peptides mimics that of eukaryotic Rce1. Its structure, comprising eight transmembrane α-helices, and catalytic site are distinct from those of other IMPs. The catalytic residues are located ∼10 Å into the membrane and are exposed to the cytoplasm and membrane through a conical cavity that accommodates the prenylated CAAX substrate. We propose that the farnesyl lipid binds to a site at the opening of two transmembrane α-helices, which results in the scissile bond being positioned adjacent to a glutamate-activated nucleophilic water molecule. This study suggests that Rce1 is the founding member of a novel IMP family, the glutamate IMPs.
- Institute of Cancer Research, 237 Fulham Road, London, SW3 6JB, UK.
Organizational Affiliation: