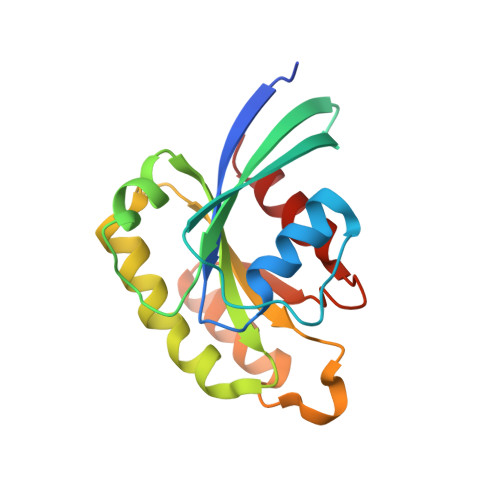
Structural and Functional Analysis of Fip2 Binding to the Endosome-Localised Rab25 Gtpase
Lall, P., Horgan, C.P., Oda, S., Franklin, E., Sultana, A., Hanscomb, S.R., Mccaffrey, M.W., Khan, A.R.(2013) Biochim Biophys Acta 1834: 2679
- PubMed: 24056041
- DOI: https://doi.org/10.1016/j.bbapap.2013.09.005
- Primary Citation of Related Structures:
4C4P - PubMed Abstract:
Rab small GTPases are the master regulators of intracellular trafficking in eukaryotes. They mediate spatial and temporal recruitment of effector proteins to distinct cellular compartments through GTP-induced changes in their conformation. Despite numerous structural studies, the molecular basis for Rab/effector specificity and subsequent biological activity remains poorly understood. Rab25, also known as Rab11c, which is epithelial-specific, has been heavily implicated in ovarian cancer development and independently appears to act as a tumour suppressor in the context of a distinct subset of carcinomas. Here, we show that Rab25 associates with FIP2 and can recruit this effector protein to endosomal membranes. We report the crystal structure of Rab25 in complex with the C-terminal region of FIP2, which consists of a central dimeric FIP2 coiled-coil that mediates a heterotetrameric Rab25-(FIP2)2-Rab25 complex. Thermodynamic analyses show that, despite a relatively conserved interface, FIP2 binds to Rab25 with an approximate 3-fold weaker affinity than to Rab11a. Reduced affinity is mainly associated with lower enthalpic gains for Rab25:FIP2 complex formation, and can be attributed to subtle differences in the conformations of switch 1 and switch 2. These cellular, structural and thermodynamic studies provide insight into the Rab11/Rab25 subfamily of small GTPases that regulate endosomal trafficking pathways in eukaryotes.
- School of Biochemistry and Immunology, Trinity College, Dublin 2, Ireland.
Organizational Affiliation: