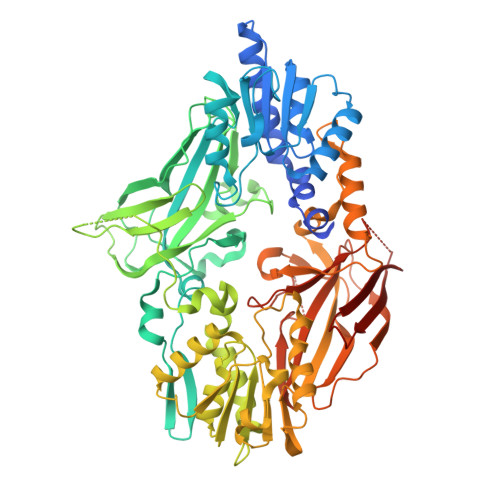
Structural Insight Into Arginine Methylation by the Mouse Protein Arginine Methyltransferase 7: A Zinc Finger Freezes the Mimic of the Dimeric State Into a Single Active Site.
Cura, V., Troffer-Charlier, N., Wurtz, J.M., Bonnefond, L., Cavarelli, J.(2014) Acta Crystallogr D Biol Crystallogr 70: 2401
- PubMed: 25195753
- DOI: https://doi.org/10.1107/S1399004714014278
- Primary Citation of Related Structures:
4C4A - PubMed Abstract:
Protein arginine methyltransferase 7 (PRMT7) is a type III arginine methyltransferase which has been implicated in several biological processes such as transcriptional regulation, DNA damage repair, RNA splicing, cell differentiation and metastasis. PRMT7 is a unique but less characterized member of the family of PRMTs. The crystal structure of full-length PRMT7 from Mus musculus refined at 1.7 Å resolution is described. The PRMT7 structure is composed of two catalytic modules in tandem forming a pseudo-dimer and contains only one AdoHcy molecule bound to the N-terminal module. The high-resolution crystal structure presented here revealed several structural features showing that the second active site is frozen in an inactive state by a conserved zinc finger located at the junction between the two PRMT modules and by the collapse of two degenerated AdoMet-binding loops.
- Département de Biologie Structurale Intégrative, Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), Université de Strasbourg, CNRS UMR7104, INSERM U596, 1 Rue Laurent Fries, F-67404 Illkirch, France.
Organizational Affiliation: