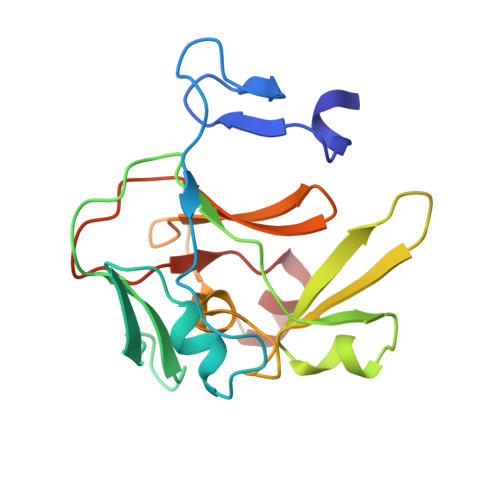
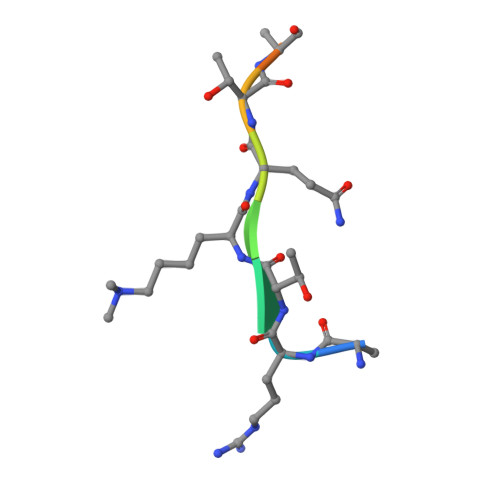
Molecular Basis for the Regulation of the H3K4 Methyltransferase Activity of Prdm9.
Wu, H., Mathioudakis, N., Diagouraga, B., Dong, A., Dombrovski, L., Baudat, F., Cusack, S., De Massy, B., Kadlec, J.(2013) Cell Rep 5: 13
- PubMed: 24095733
- DOI: https://doi.org/10.1016/j.celrep.2013.08.035
- Primary Citation of Related Structures:
4C1Q - PubMed Abstract:
PRDM9, a histone lysine methyltransferase, is a key determinant of the localization of meiotic recombination hot spots in humans and mice and the only vertebrate protein known to be involved in hybrid sterility. Here, we report the crystal structure of the PRDM9 methyltransferase domain in complex with a histone H3 peptide dimethylated on lysine 4 (H3K4me2) and S-adenosylhomocysteine (AdoHcy), which provides insights into the methyltransferase activity of PRDM proteins. We show that the genuine substrate of PRDM9 is histone H3 lysine 4 (H3K4) and that the enzyme possesses mono-, di-, and trimethylation activities. We also determined the crystal structure of PRDM9 in its autoinhibited state, which revealed a rearrangement of the substrate and cofactor binding sites by a concerted action of the pre-SET and post-SET domains, providing important insights into the regulatory mechanisms of histone lysine methyltransferase activity.
- Structural Genomics Consortium, University of Toronto, MaRS Center, South Tower, 7(th) Floor, 101 College Street, Toronto, ON M5G 1L7, Canada.
Organizational Affiliation: