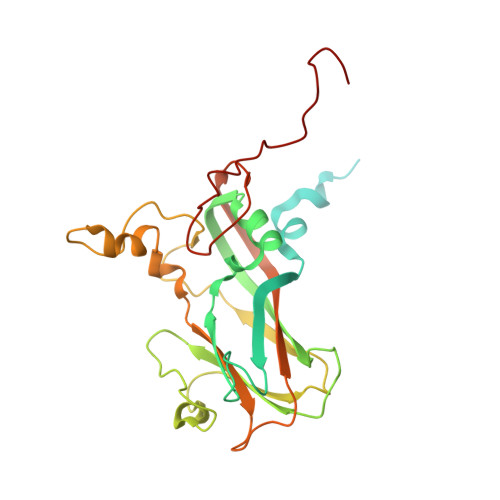
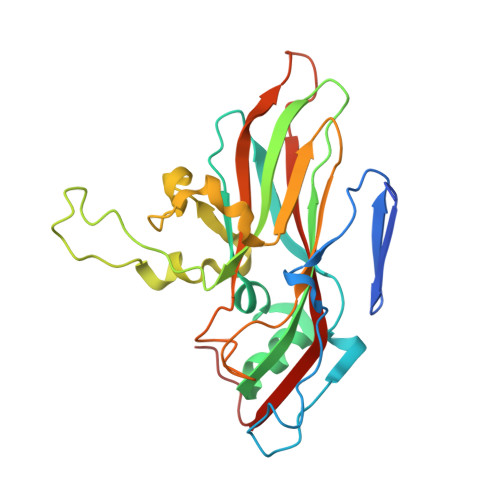
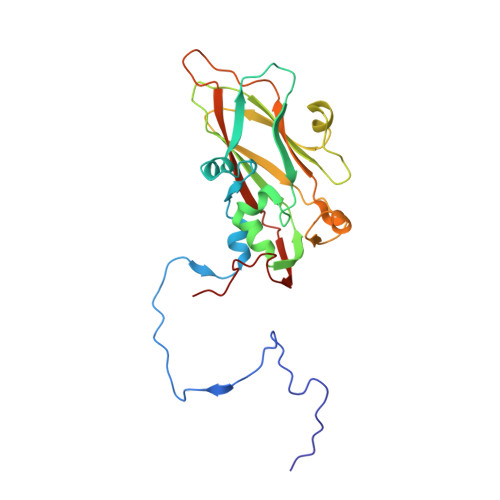
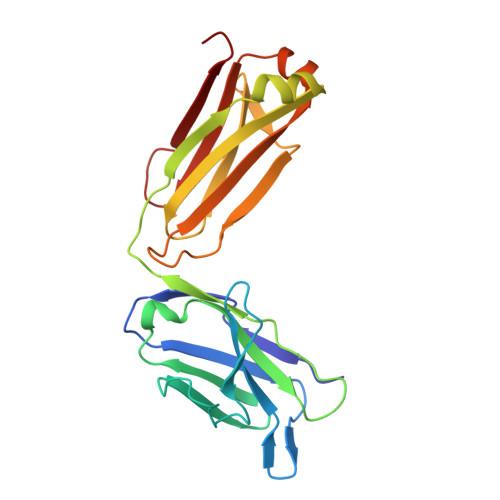
Neutralizing Antibodies Can Initiate Genome Release from Human Enterovirus 71.
Plevka, P., Lim, P., Perera, R., Cardosa, J., Suksatu, A., Kuhn, R.J., Rossmann, M.G.(2014) Proc Natl Acad Sci U S A 111: 2134
- PubMed: 24469789
- DOI: https://doi.org/10.1073/pnas.1320624111
- Primary Citation of Related Structures:
4C0U, 4C0Y, 4C10 - PubMed Abstract:
Antibodies were prepared by immunizing mice with empty, immature particles of human enterovirus 71 (EV71), a picornavirus that causes severe neurological disease in young children. The capsid structure of these empty particles is different from that of the mature virus and is similar to "A" particles encountered when picornaviruses recognize a potential host cell before genome release. The monoclonal antibody E18, generated by this immunization, induced a conformational change when incubated at temperatures between 4 °C and 37 °C with mature virus, transforming infectious virions into A particles. The resultant loss of genome that was observed by cryo-EM and a fluorescent SYBR Green dye assay inactivated the virus, establishing the mechanism by which the virus is inactivated and demonstrating that the E18 antibody has potential as an anti-EV71 therapy. The antibody-mediated virus neutralization by the induction of genome release has not been previously demonstrated. Furthermore, the present results indicate that antibodies with genome-release activity could also be produced for other picornaviruses by immunization with immature particles.
- Department of Biological Sciences, Purdue University, West Lafayette, IN 47907.
Organizational Affiliation: