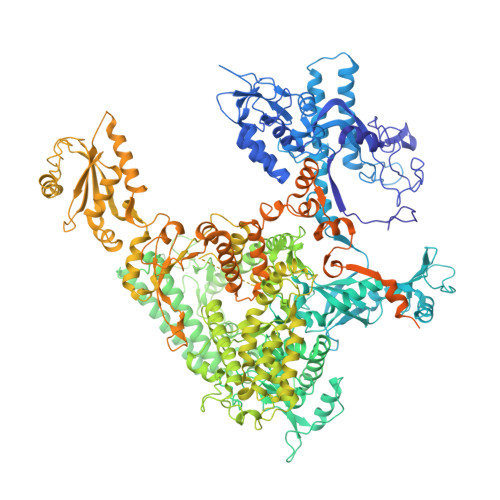
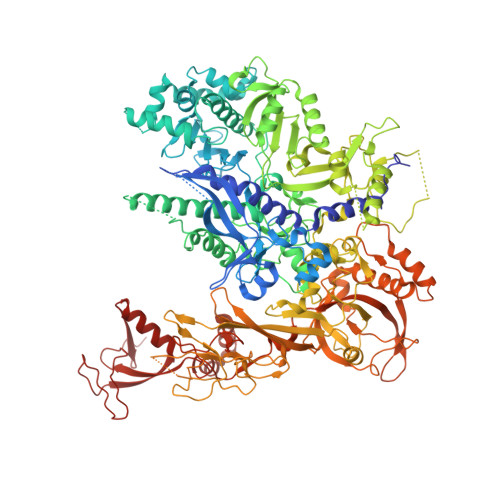
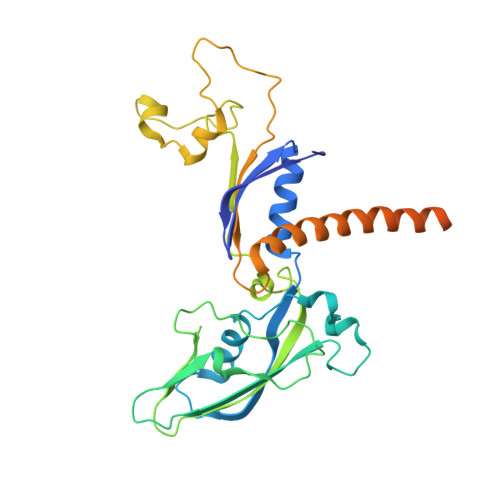
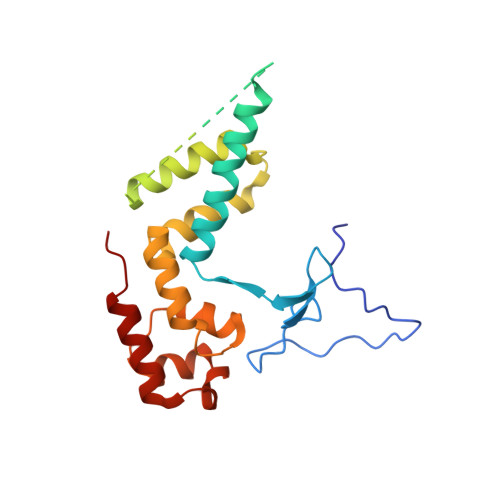
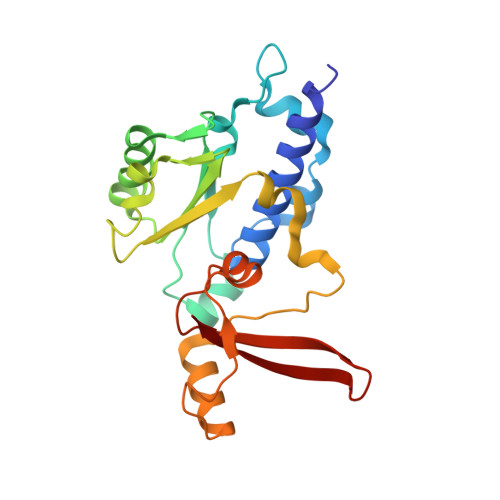
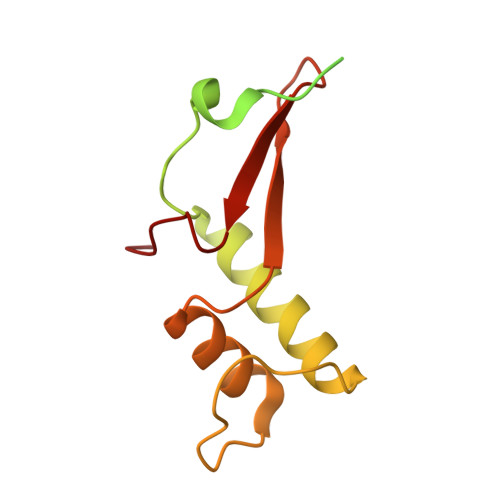
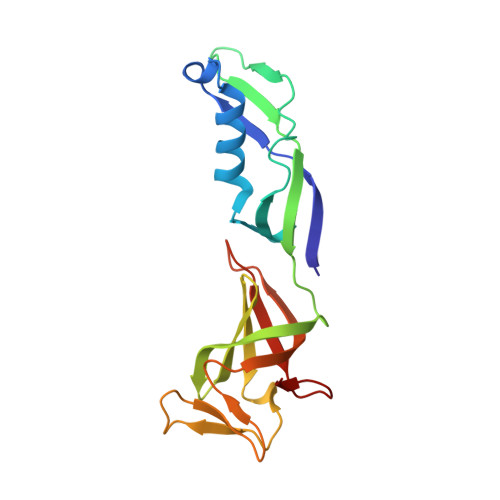
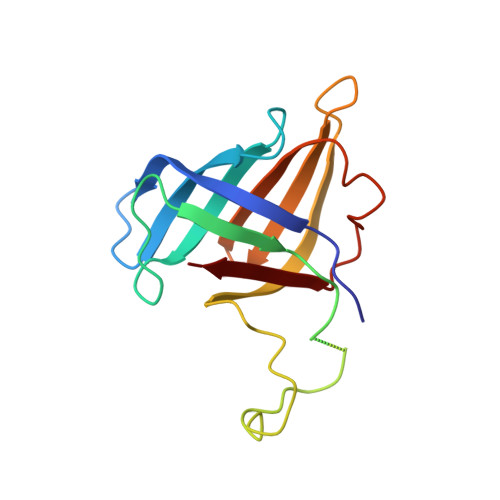
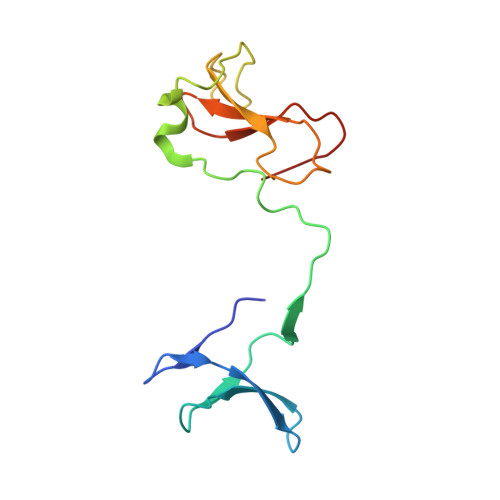
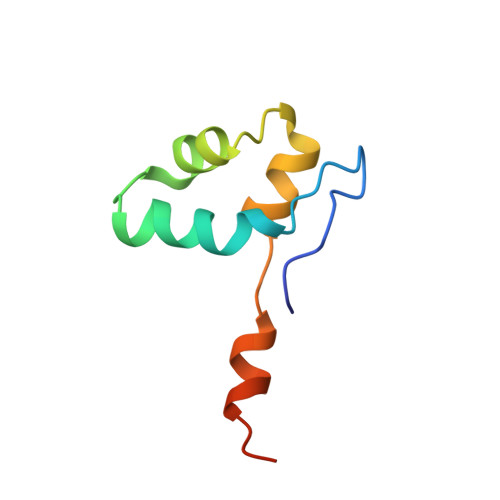
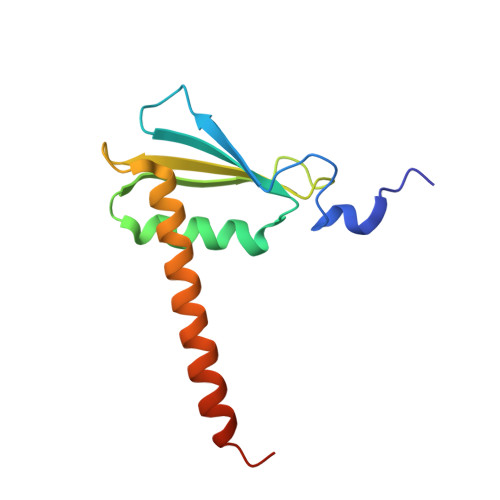
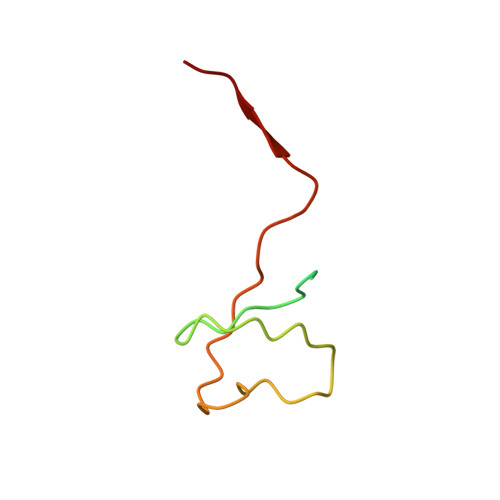
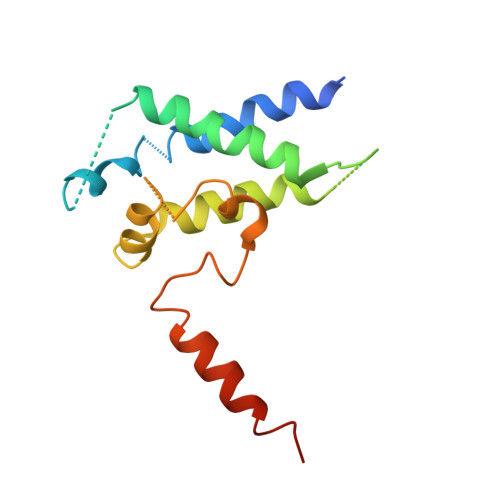
Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Kinkelin, K., Wozniak, G.G., Rothbart, S.B., Lidschreiber, M., Strahl, B.D., Cramer, P.(2013) Proc Natl Acad Sci U S A 110: 15277
- PubMed: 24003114
- DOI: https://doi.org/10.1073/pnas.1311010110
- Primary Citation of Related Structures:
4BXX, 4BXZ, 4BY1, 4BY7 - PubMed Abstract:
Bypass of Ess1 (Bye1) is a nuclear protein with a domain resembling the central domain in the transcription elongation factor TFIIS. Here we show that Bye1 binds with its TFIIS-like domain (TLD) to RNA polymerase (Pol) II, and report crystal structures of the Bye1 TLD bound to Pol II and three different Pol II-nucleic acid complexes. Like TFIIS, Bye1 binds with its TLD to the Pol II jaw and funnel. In contrast to TFIIS, however, it neither alters the conformation nor the in vitro functions of Pol II. In vivo, Bye1 is recruited to chromatin via its TLD and occupies the 5'-region of active genes. A plant homeo domain (PHD) in Bye1 binds histone H3 tails with trimethylated lysine 4, and this interaction is enhanced by the presence of neighboring posttranslational modifications (PTMs) that mark active transcription and conversely is impaired by repressive PTMs. We identify putative human homologs of Bye1, the proteins PHD finger protein 3 and death-inducer obliterator, which are both implicated in cancer. These results establish Bye1 as the founding member of a unique family of chromatin transcription factors that link histones with active PTMs to transcribing Pol II.
- Gene Center Munich and Department of Biochemistry, Center for Integrated Protein Science Munich (CIPSM), Ludwig-Maximilians-Universität München, 81377 Munich, Germany.
Organizational Affiliation: