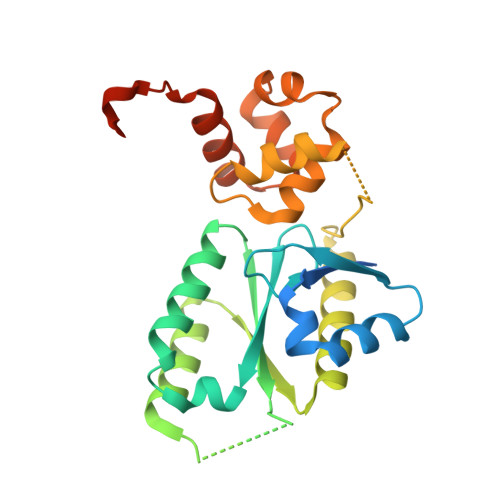
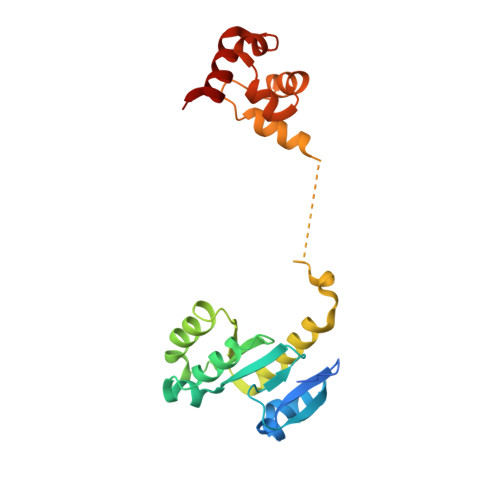
Architecture and DNA Recognition Elements of the Fanconi Anemia Fancm-Faap24 Complex.
Coulthard, R., Deans, A.J., Swuec, P., Bowles, M., Costa, A., West, S.C., Mcdonald, N.Q.(2013) Structure 21: 1648
- PubMed: 23932590
- DOI: https://doi.org/10.1016/j.str.2013.07.006
- Primary Citation of Related Structures:
4BXO - PubMed Abstract:
Fanconi anemia (FA) is a disorder associated with a failure in DNA repair. FANCM (defective in FA complementation group M) and its partner FAAP24 target other FA proteins to sites of DNA damage. FANCM-FAAP24 is related to XPF/MUS81 endonucleases but lacks endonucleolytic activity. We report a structure of an FANCM C-terminal fragment (FANCMCTD) bound to FAAP24 and DNA. This S-shaped structure reveals the FANCM (HhH)2 domain is buried, whereas the FAAP24 (HhH)2 domain engages DNA. We identify a second DNA contact and a metal center within the FANCM pseudo-nuclease domain and demonstrate that mutations in either region impair double-stranded DNA binding in vitro and FANCM-FAAP24 function in vivo. We show the FANCM translocase domain lies in proximity to FANCMCTD by electron microscopy and that binding fork DNA structures stimulate its ATPase activity. This suggests a tracking model for FANCM-FAAP24 until an encounter with a stalled replication fork triggers ATPase-mediated fork remodeling.
- Structural Biology Laboratory, Cancer Research UK, London Research Institute, 44 Lincoln's Inn Fields, London WC2A 3LY, UK.
Organizational Affiliation: