Structure of the Cyanobacterial Phytochrome 2 Photosensor Implies a Tryptophan Switch for Phytochrome Signaling.
Anders, K., Daminelli-Widany, G., Mroginski, M.A., von Stetten, D., Essen, L.-O.(2013) J Biological Chem 288: 35714
- PubMed: 24174528
- DOI: https://doi.org/10.1074/jbc.M113.510461
- Primary Citation of Related Structures:
4BWI - PubMed Abstract:
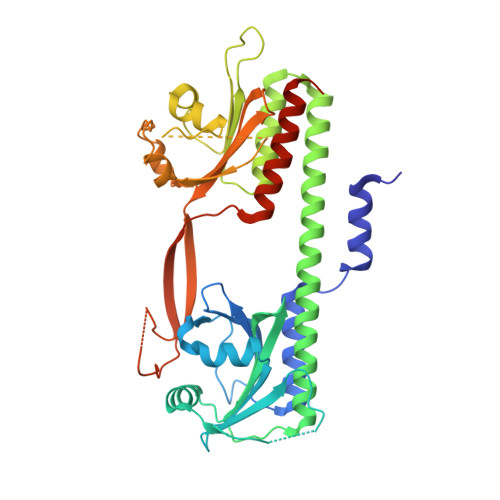
Phytochromes are highly versatile photoreceptors, which occur ubiquitously in plants as well as in many light-responsive microorganisms. Here, photosynthetic cyanobacteria utilize up to three different phytochrome architectures, where only the plant-like and the single-domain cyanobacteriochromes are structurally characterized so far. Cph2 represents a third group in Synechocystis species and affects their capability of phototaxis by controlling c-di-GMP synthesis and degradation. The 2.6-Å crystal structure of its red/far-red responsive photosensory module in the Pr state reveals a tandem-GAF bidomain that lacks the figure-of-eight knot of the plant/cph1 subfamily. Its covalently attached phycocyanobilin chromophore adopts a highly tilted ZZZssa conformation with a novel set of interactions between its propionates and the GAF1 domain. The tongue-like protrusion from the GAF2 domain interacts with the GAF1-bound chromophore via its conserved PRXSF, WXE, and W(G/A)G motifs. Mutagenesis showed that the integrity of the tongue is indispensable for Pr → Pfr photoconversion and involves a swap of the motifs' tryptophans within the tongue-GAF1 interface. This "Trp switch" is supposed to be a crucial element for the photochromicity of all multidomain phytochromes.
- From the Department of Chemistry, Biomedical Research Centre, Philipps-Universität, D-35032 Marburg, Germany.
Organizational Affiliation: