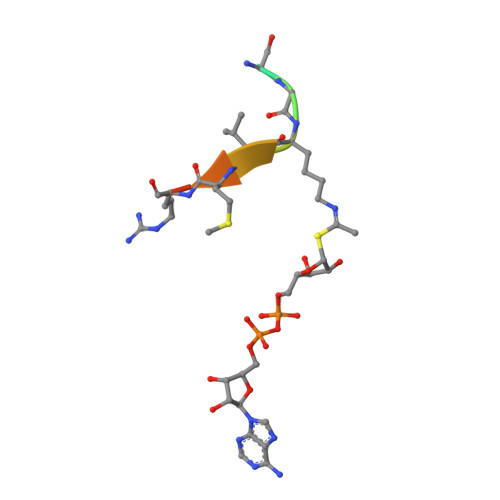
Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Gertz, M., Fischer, F., Nguyen, G.T.T., Lakshminarasimhan, M., Schutkowski, M., Weyand, M., Steegborn, C.(2013) Proc Natl Acad Sci U S A 110: E2772
- PubMed: 23840057
- DOI: https://doi.org/10.1073/pnas.1303628110
- Primary Citation of Related Structures:
4BUZ, 4BV2, 4BV3, 4BVB, 4BVE, 4BVF, 4BVG, 4BVH - PubMed Abstract:
Sirtuins are protein deacetylases regulating metabolism and stress responses. The seven human Sirtuins (Sirt1-7) are attractive drug targets, but Sirtuin inhibition mechanisms are mostly unidentified. We report the molecular mechanism of Sirtuin inhibition by 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide (Ex-527). Inhibitor binding to potently inhibited Sirt1 and Thermotoga maritima Sir2 and to moderately inhibited Sirt3 requires NAD(+), alone or together with acetylpeptide. Crystal structures of several Sirtuin inhibitor complexes show that Ex-527 occupies the nicotinamide site and a neighboring pocket and contacts the ribose of NAD(+) or of the coproduct 2'-O-acetyl-ADP ribose. Complex structures with native alkylimidate and thio-analog support its catalytic relevance and show, together with biochemical assays, that only the coproduct complex is relevant for inhibition by Ex-527, which stabilizes the closed enzyme conformation preventing product release. Ex-527 inhibition thus exploits Sirtuin catalysis, and kinetic isoform differences explain its selectivity. Our results provide insights in Sirtuin catalysis and inhibition with important implications for drug development.
- Department of Biochemistry and Research Center for Bio-Macromolecules, University of Bayreuth, 95440 Bayreuth, Germany.
Organizational Affiliation: