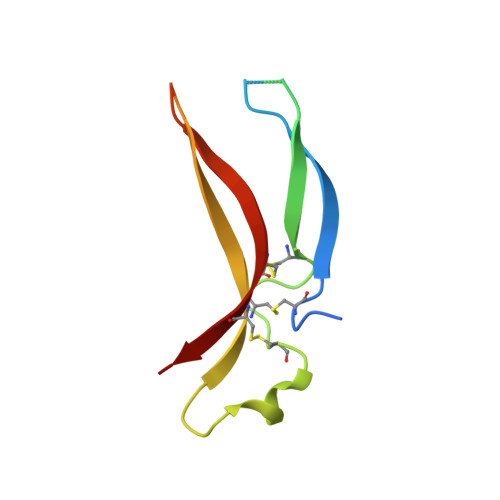
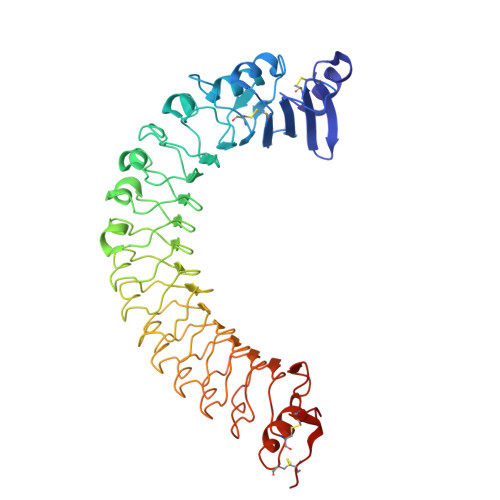
Cytokine Spatzle Binds to the Drosophila Immunoreceptor Toll with a Neurotrophin-Like Specificity and Couples Receptor Activation.
Lewis, M., Arnot, C.J., Beeston, H., Mccoy, A., Ashcroft, A.E., Gay, N.J., Gangloff, M.(2013) Proc Natl Acad Sci U S A 110: 20461
- PubMed: 24282309
- DOI: https://doi.org/10.1073/pnas.1317002110
- Primary Citation of Related Structures:
4BV4 - PubMed Abstract:
Drosophila Toll functions in embryonic development and innate immunity and is activated by an endogenous ligand, Spätzle (Spz). The related Toll-like receptors in vertebrates also function in immunity but are activated directly by pathogen-associated molecules such as bacterial endotoxin. Here, we present the crystal structure at 2.35-Å resolution of dimeric Spz bound to a Toll ectodomain encompassing the first 13 leucine-rich repeats. The cystine knot of Spz binds the concave face of the Toll leucine-rich repeat solenoid in an area delineated by N-linked glycans and induces a conformational change. Mutagenesis studies confirm that the interface observed in the crystal structure is relevant for signaling. The asymmetric binding mode of Spz to Toll is similar to that of nerve growth factor (NGF) in complex with the p75 neurotrophin receptor but is distinct from that of microbial ligands bound to the Toll-like receptors. Overall, this study indicates an allosteric signaling mechanism for Toll in which ligand binding to the N terminus induces a conformational change that couples to homodimerization of juxtamembrane structures in the Toll ectodomain C terminus.
- Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, United Kingdom.
Organizational Affiliation: