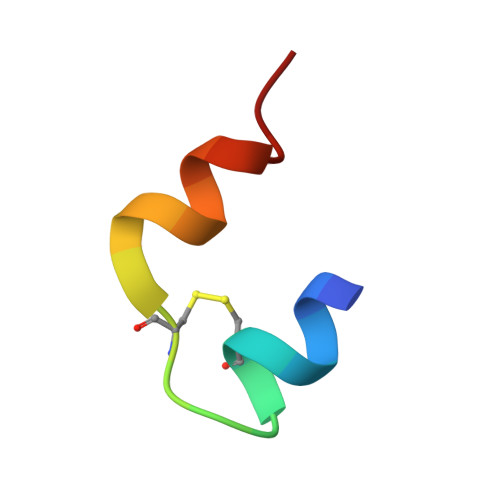
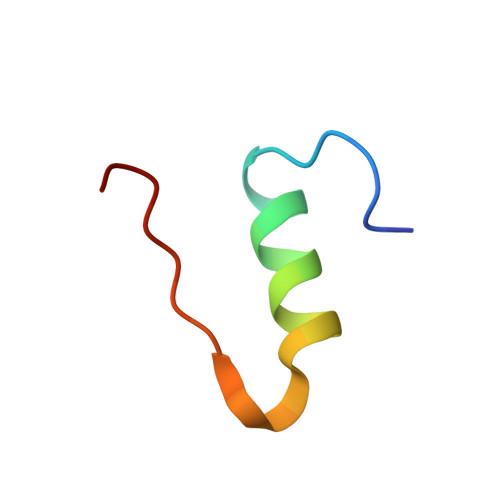
Chipx: A Novel Microfluidic Chip for Counter- Diffusion Crystallization of Biomolecules and in Situ Crystal Analysis at Room Temperature
Pinker, F., Brun, M., Morin, P., Deman, A.-L., Chateau, J.-F., Olieric, V., Stirnimann, C., Lorber, B., Terrier, N., Ferrigno, R., Sauter, C.(2013) J Cryst Growth 13: 3333