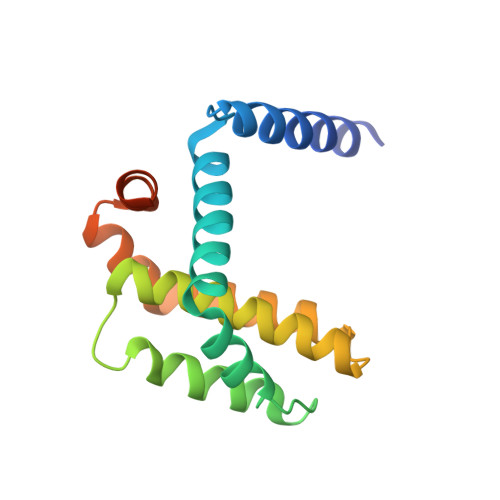
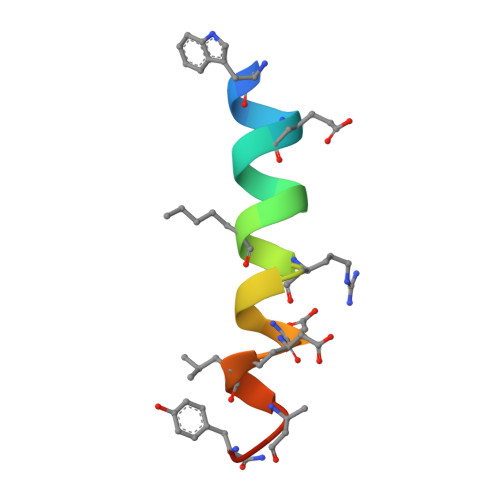
Structure-Guided Rational Design of Alpha/Beta-Peptide Foldamers with High Affinity for Bcl-2 Family Prosurvival Proteins.
Smith, B.J., Lee, E.F., Checco, J.W., Evangelista, M., Gellman, S.H., Fairlie, W.D.(2013) Chembiochem 14: 1564
- PubMed: 23929624
- DOI: https://doi.org/10.1002/cbic.201300351
- Primary Citation of Related Structures:
4BPI, 4BPJ, 4BPK - PubMed Abstract:
We have used computational methods to improve the affinity of a foldamer ligand for its target protein. The effort began with a previously reported α/β-peptide based on the BH3 domain of the proapoptotic protein Puma; this foldamer binds tightly to Bcl-x(L) but weakly to Mcl-1. The crystal structure of the Puma-derived α/β-peptide complexed to Bcl-x(L) was used as the basis for computational design of variants intended to display improved binding to Mcl-1. Molecular modelling suggested modification of three α residues of the original α/β backbone. Individually, each substitution caused only a modest (4- to 15-fold) gain in affinity; however, together the three substitutions led to a 250-fold increase in binding to Mcl-1. These modifications had very little effect on affinity for Bcl-x(L). Crystal structures of a number of the new α/β-peptides bound to either Mcl-1 or Bcl-x(L) validated the selection of each substitution. Overall, our findings demonstrate that structure-guided rational design can be used to improve affinity and alter partner selectivity of peptidic ligands with unnatural backbones that bind to specific protein partners.
- Department of Chemistry, La Trobe Institute for Molecular Science, La Trobe University, Bundoora, Victoria (Australia). brian.smith@latrobe.edu.au.
Organizational Affiliation: