Structural Characterizations of Chloroplast Translocon Protein Tic110.
Tsai, J.-Y., Chu, C.-C., Yeh, Y.-H., Chen, L.-J., Li, H.-M., Hsiao, C.-D.(2013) Plant J 75: 847
- PubMed: 23711301
- DOI: https://doi.org/10.1111/tpj.12249
- Primary Citation of Related Structures:
4BM5 - PubMed Abstract:
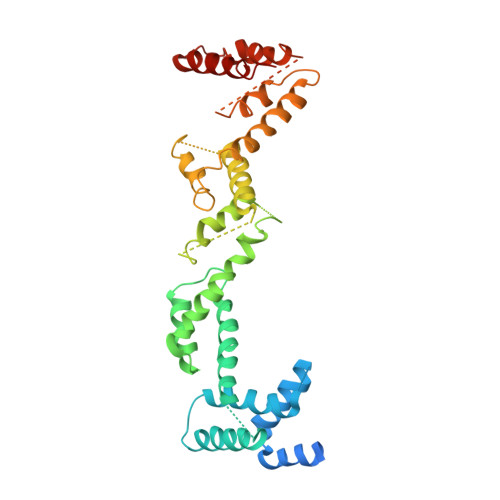
Tic110 is a major component of the chloroplast protein import translocon. Two functions with mutually exclusive structures have been proposed for Tic110: a protein-conducting channel with six transmembrane domains and a scaffold with two N-terminal transmembrane domains followed by a large soluble domain for binding transit peptides and other stromal translocon components. To investigate the structure of Tic110, Tic110 from Cyanidioschyzon merolae (CmTic110) was characterized. We constructed three fragments, CmTic110A , CmTic110B and CmTic110C , with increasing N-terminal truncations, to perform small-angle X-ray scattering (SAXS) and X-ray crystallography analyses and Dali structural comparison. Here we report the molecular envelope of CmTic110B and CmTic110C determined by SAXS, and the crystal structure of CmTic110C at 4.2 Å. Our data indicate that the C-terminal half of CmTic110 possesses a rod-shaped helix-repeat structure that is too flattened and elongated to be a channel. The structure is most similar to the HEAT-repeat motif that functions as scaffolds for protein-protein interactions.
- Institute of Molecular Biology, Academia Sinica, Taipei, 11529, Taiwan.
Organizational Affiliation: