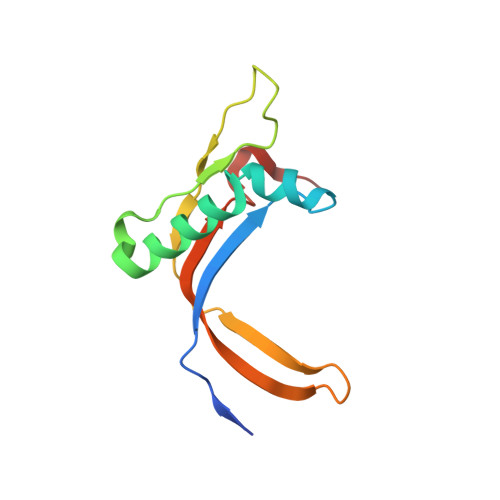
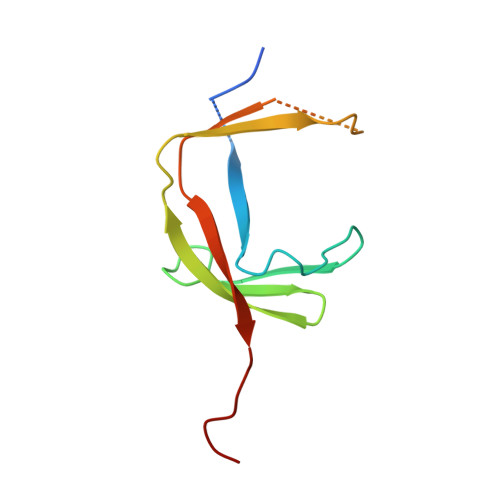
RNA Polymerase III-Specific General Transcription Factor Iiic Contains a Heterodimer Resembling Tfiif RAP30/RAP74.
Taylor, N.M.I., Baudin, F., Von Scheven, G., Muller, C.W.(2013) Nucleic Acids Res 41: 9183
- PubMed: 23921640
- DOI: https://doi.org/10.1093/nar/gkt664
- Primary Citation of Related Structures:
4BJI, 4BJJ - PubMed Abstract:
Transcription of tRNA-encoding genes by RNA polymerase (Pol) III requires the six-subunit general transcription factor IIIC that uses subcomplexes τA and τB to recognize two gene-internal promoter elements named A- and B-box. The Schizosaccharomyces pombe τA subcomplex comprises subunits Sfc1, Sfc4 and Sfc7. The crystal structure of the Sfc1/Sfc7 heterodimer reveals similar domains and overall domain architecture to the Pol II-specific general transcription factor TFIIF Rap30/Rap74. The N-terminal Sfc1/Sfc7 dimerization module consists of a triple β-barrel similar to the N-terminal TFIIF Rap30/Rap74 dimerization module, whereas the C-terminal Sfc1 DNA-binding domain contains a winged-helix domain most similar to the TFIIF Rap30 C-terminal winged-helix domain. Sfc1 DNA-binding domain recognizes single and double-stranded DNA by an unknown mechanism. Several features observed for A-box recognition by τA resemble the recognition of promoters by bacterial RNA polymerase, where σ factor unfolds double-stranded DNA and stabilizes the non-coding DNA strand in an open conformation. Such a function has also been proposed for TFIIF, suggesting that the observed structural similarity between Sfc1/Sfc7 and TFIIF Rap30/Rap74 might also reflect similar functions.
- European Molecular Biology Laboratory (EMBL), Structural and Computational Biology Unit, Meyerhofstrasse 1, 69117 Heidelberg, Germany and UJF-EMBL-CNRS UMI 3265, Unit of Virus Host-Cell Interactions, 38042 Grenoble Cedex 9, France.
Organizational Affiliation: