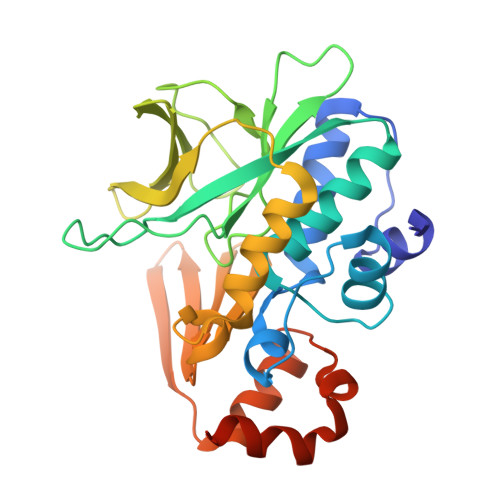
Structure of arylamine N-acetyltransferase from Mycobacterium tuberculosis determined by cross-seeding with the homologous protein from M. marinum: triumph over adversity.
Abuhammad, A., Lowe, E.D., McDonough, M.A., Shaw Stewart, P.D., Kolek, S.A., Sim, E., Garman, E.F.(2013) Acta Crystallogr D Biol Crystallogr 69: 1433-1446
- PubMed: 23897467
- DOI: https://doi.org/10.1107/S0907444913015126
- Primary Citation of Related Structures:
4BGF - PubMed Abstract:
Arylamine N-acetyltransferase from Mycobacterium tuberculosis (TBNAT) plays an important role in the intracellular survival of the microorganism inside macrophages. Medicinal chemistry efforts to optimize inhibitors of the TBNAT enzyme have been hampered by the lack of a three-dimensional structure of the enzyme. In this paper, the first structure of TBNAT, determined using a lone crystal produced using cross-seeding with the homologous protein from M. marinum, is reported. Despite the similarity between the two enzymes (74% sequence identity), they show distinct physical and biochemical characteristics. The structure elegantly reveals the characteristic features of the protein surface as well as details of the active site of TBNAT relevant to drug-discovery efforts. The crystallographic analysis of the diffraction data presented many challenges, since the crystal was twinned and the habit possessed pseudo-translational symmetry.
- Department of Pharmacology, University of Oxford, Mansfield Road, Oxford OX1 3QT, England.
Organizational Affiliation: