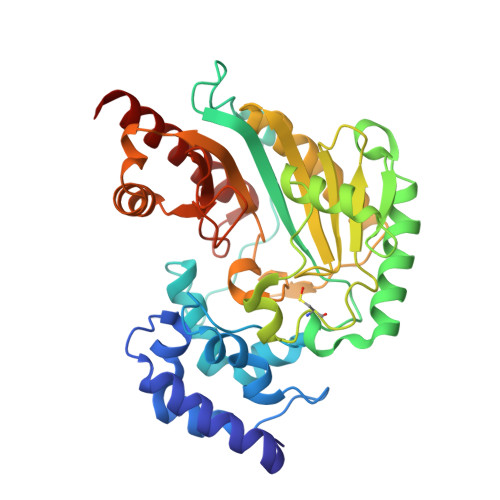
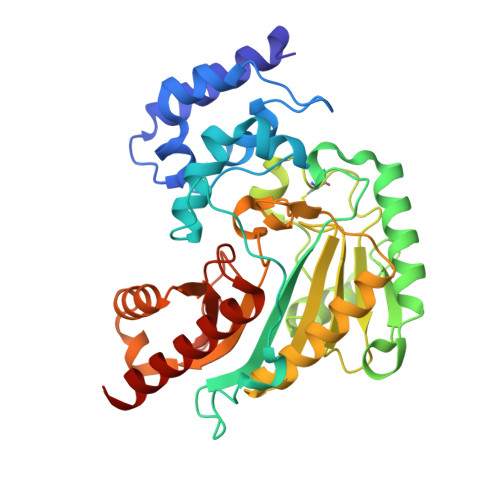
Crystal Structure of Shrimp Arginine Kinase in Binary Complex with Arginine-A Molecular View of the Phosphagen Precursor Binding to the Enzyme.
Lopez-Zavala, A.A., Garcia-Orozco, K.D., Carrasco-Miranda, J.S., Sugich-Miranda, R., Velazquez-Contreras, E.F., Criscitiello, M.F., Brieba, L.G., Rudino-Pinera, E., Sotelo-Mundo, R.R.(2013) J Bioenerg Biomembr 45: 511
- PubMed: 23873077
- DOI: https://doi.org/10.1007/s10863-013-9521-0
- Primary Citation of Related Structures:
4BG4, 4BHL - PubMed Abstract:
Arginine kinase (AK) is a key enzyme for energetic balance in invertebrates. Although AK is a well-studied system that provides fast energy to invertebrates using the phosphagen phospho-arginine, the structural details on the AK-arginine binary complex interaction remain unclear. Herein, we determined two crystal structures of the Pacific whiteleg shrimp (Litopenaeus vannamei) arginine kinase, one in binary complex with arginine (LvAK-Arg) and a ternary transition state analog complex (TSAC). We found that the arginine guanidinium group makes ionic contacts with Glu225, Cys271 and a network of ordered water molecules. On the zwitterionic side of the amino acid, the backbone amide nitrogens of Gly64 and Val65 coordinate the arginine carboxylate. Glu314, one of proposed acid-base catalytic residues, did not interact with arginine in the binary complex. This residue is located in the flexible loop 310-320 that covers the active site and only stabilizes in the LvAK-TSAC. This is the first binary complex crystal structure of a guanidine kinase in complex with the guanidine substrate and could give insights into the nature of the early steps of phosphagen biosynthesis.
- Centro de Investigación en Alimentación y Desarrollo, A.C. (CIAD). Carretera a Ejido La Victoria Km 0.6, Apartado Postal 1735, Hermosillo, Sonora, 83304, Mexico.
Organizational Affiliation: