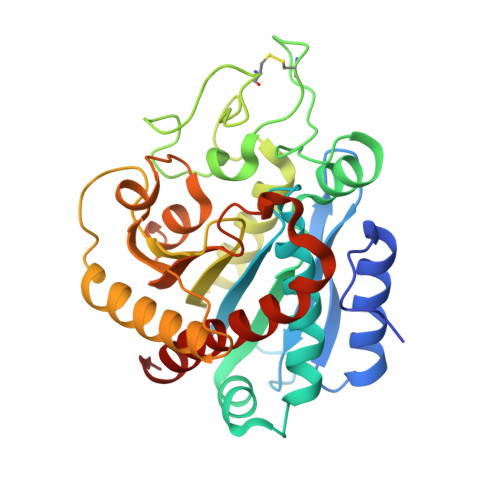
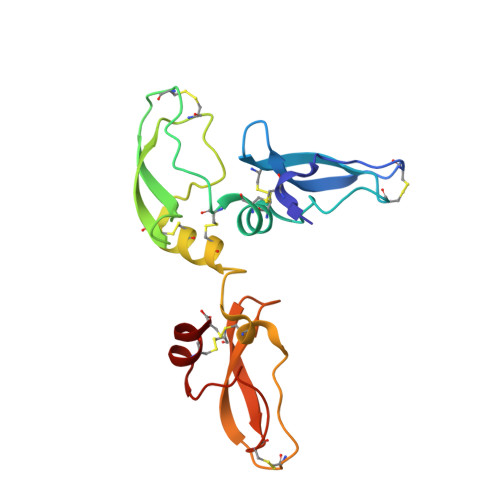
A Noncanonical Mechanism of Carboxypeptidase Inhibition Revealed by the Crystal Structure of the Tri-Kunitz Smci in Complex with Human Cpa4.
Alonso Del Rivero, M., Reytor, M.L., Trejo, S.A., Chavez, M.A., Aviles, F.X., Reverter, D.(2013) Structure 21: 1118
- PubMed: 23746805
- DOI: https://doi.org/10.1016/j.str.2013.04.021
- Primary Citation of Related Structures:
4BD9 - PubMed Abstract:
The crystal structure of SmCI (Sabellastarte magnifica carboxypeptidase inhibitor) has been determined in complex with human carboxypeptidase A4 (hCPA4). SmCI is composed by three BPTI/Kunitz domains, each one displaying high structural homology and functionality with serine protease inhibitors. Moreover, SmCI possesses a distinctive capability to inhibit metallo-carboxypeptidases, constituting a bifunctional metallocarboxy- and serine protease inhibitor. The structure of the 1:1 complex of SmCI with hCPA4 reveals a noncanonical mechanism of carboxypeptidase inhibition, which surprisingly occurs mainly via the N-terminal segment, which penetrates into the active site groove of the enzyme. Mutagenesis and biochemical analysis confirm the major role of the N-terminal segment in the inhibition of carboxypeptidases. SmCI represents a tri-Kunitz serine protease inhibitor adapted to inhibit metallo-carboxypeptidases and discloses an unusual mechanism of inhibition by the N-terminal segment, emulating the "classical" C-terminal substrate-like inhibition.
- Centro de Estudio de Proteínas, Facultad de Biología, Universidad de la Habana, Cuba 10400.
Organizational Affiliation: