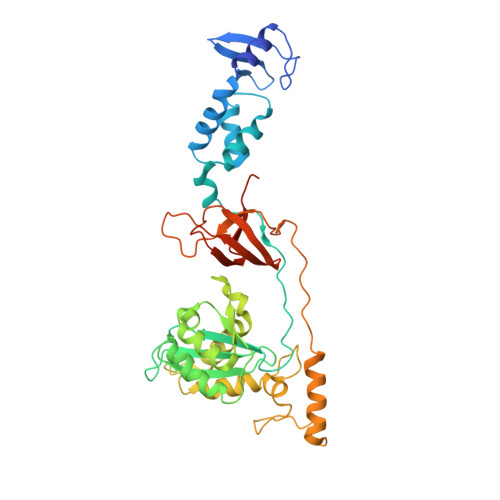
Assembly of Prototype Foamy Virus Strand Transfer Complexes on Product DNA Bypassing Catalysis of Integration.
Yin, Z., Lapkouski, M., Yang, W., Craigie, R.(2012) Protein Sci 21: 1849
- PubMed: 23011895
- DOI: https://doi.org/10.1002/pro.2166
- Primary Citation of Related Structures:
4BAC - PubMed Abstract:
Integrase is the key enzyme that mediates integration of retroviral DNA into cellular DNA which is essential for viral replication. Inhibitors of HIV-1 that target integrase recognize the nucleoprotein complexes formed by integrase and viral DNA substrate (intasomes) rather than the free enzyme. Atomic resolution structures of HIV-1 intasomes are therefore required to understand the mechanisms of inhibition and drug resistance. To date, prototype foamy virus (PFV) is the only retrovirus for which such structures have been determined. We show that PFV strand transfer complexes (STC) can be assembled on product DNA without going through the normal forward reaction pathway. The finding that a retroviral STC can be assembled in this way may provide a powerful tool to alleviate the obstacles that impede structural studies of nucleoprotein intermediates in HIV-1 DNA integration.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892, USA.
Organizational Affiliation: