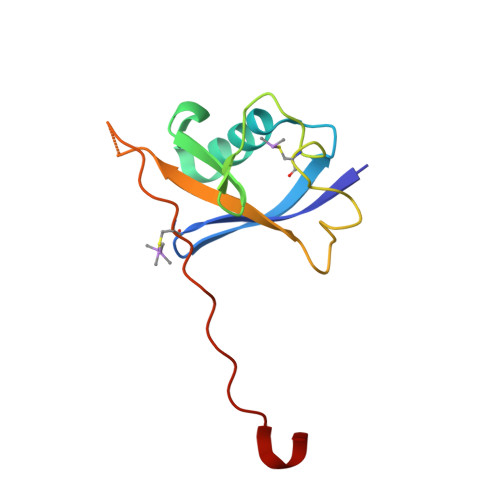
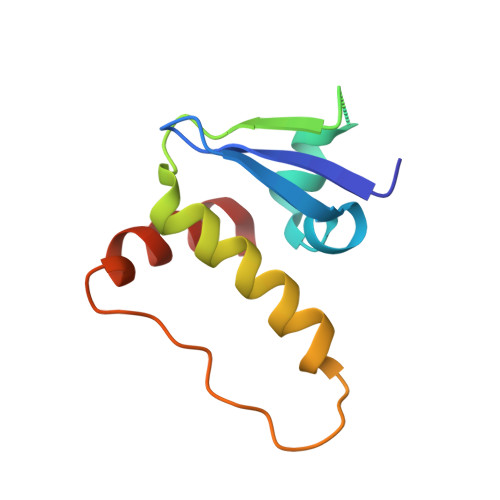
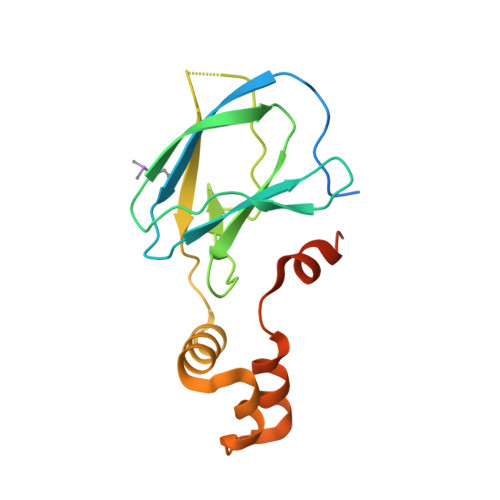
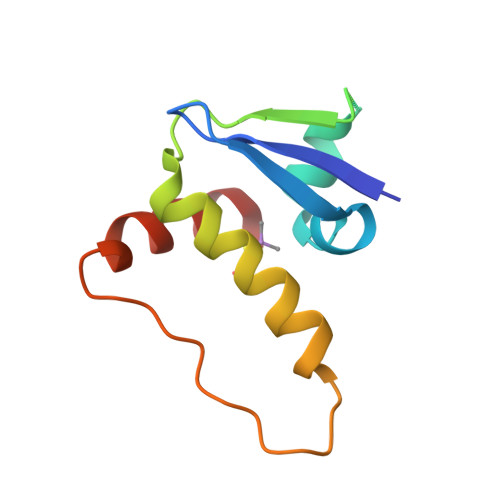
Small-Molecule Inhibitors of the Interaction between the E3 Ligase Vhl and Hif1 Alpha
Buckley, D.L., Gustafson, J.L., Van Molle, I., Roth, A.G., Tae, H.S., Gareiss, P.C., Jorgensen, W.L., Ciulli, A., Crews, C.M.(2012) Angew Chem Int Ed Engl 51: 11463
- PubMed: 23065727
- DOI: https://doi.org/10.1002/anie.201206231
- Primary Citation of Related Structures:
4B95, 4B9K - PubMed Abstract:
E3 ubiquitin ligases, such as the therapeutically relevant VHL, are challenging targets for traditional medicinal chemistry, as their modulation requires targeting protein-protein interactions. We report novel small-molecule inhibitors of the interaction between VHL and its molecular target HIF1α, a transcription factor involved in oxygen sensing.
- Department of Chemistry, Yale University, New Haven, CT 06511, USA.
Organizational Affiliation: