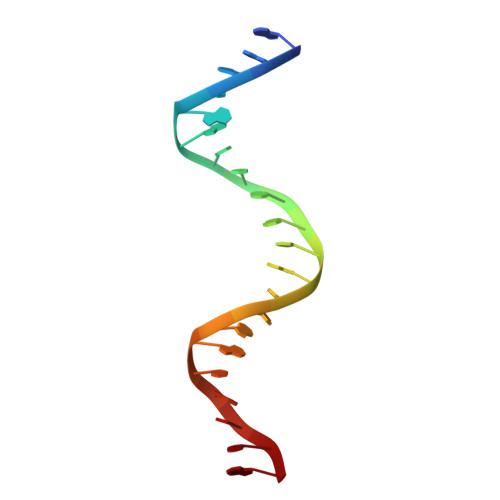
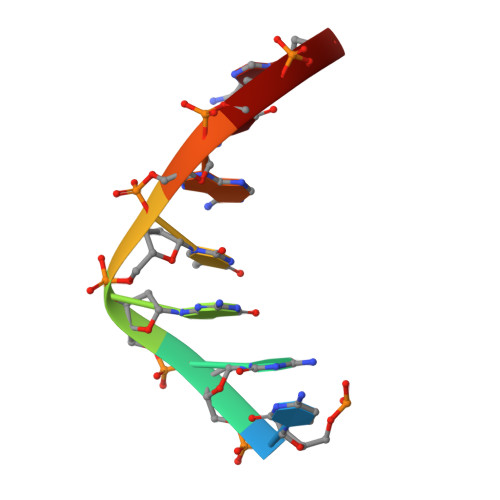
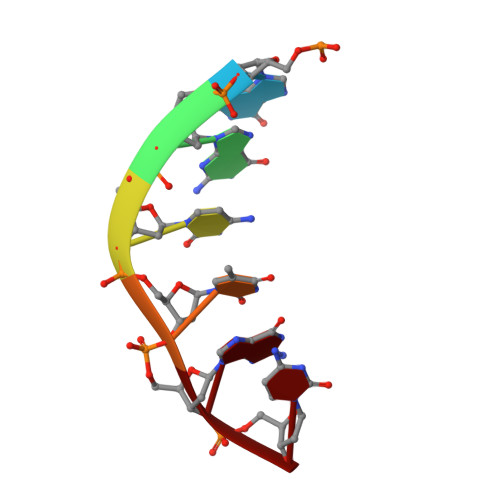
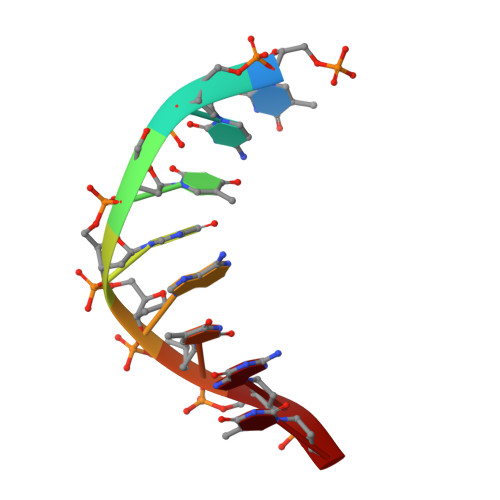
Enzymatic Production of 'Monoclonal Stoichiometric' Single-Stranded DNA Oligonucleotides
Ducani, C., Kaul, C.D., Moche, M., Shih, W.M., Hogberg, B.(2013) Nat Methods 10: 647
- PubMed: 23727986
- DOI: https://doi.org/10.1038/nmeth.2503
- Primary Citation of Related Structures:
4B8D - PubMed Abstract:
Single-stranded oligonucleotides are important as research tools, as diagnostic probes, in gene therapy and in DNA nanotechnology. Oligonucleotides are typically produced via solid-phase synthesis, using polymer chemistries that are limited relative to what biological systems produce. The number of errors in synthetic DNA increases with oligonucleotide length, and the resulting diversity of sequences can be a problem. Here we present the 'monoclonal stoichiometric' (MOSIC) method for enzyme-mediated production of DNA oligonucleotides. We amplified oligonucleotides from clonal templates derived from single bacterial colonies and then digested cutter hairpins in the products, which released pools of oligonucleotides with precisely controlled relative stoichiometric ratios. We prepared 14-378-nucleotide MOSIC oligonucleotides either by in vitro rolling-circle amplification or by amplification of phagemid DNA in Escherichia coli. Analyses of the formation of a DNA crystal and folding of DNA nanostructures confirmed the scalability, purity and stoichiometry of the produced oligonucleotides.
- Swedish Medical Nanoscience Center, Department of Neuroscience, Karolinska Institutet, Stockholm, Sweden.
Organizational Affiliation: