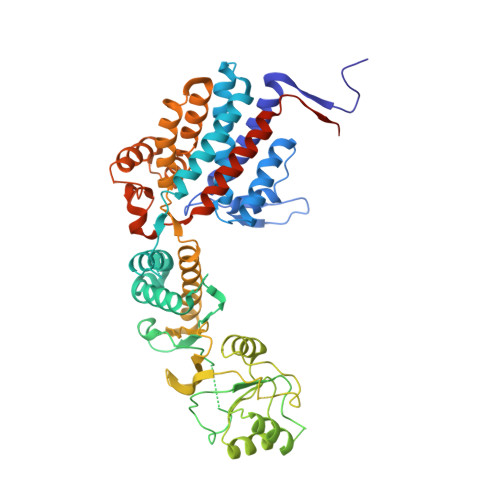
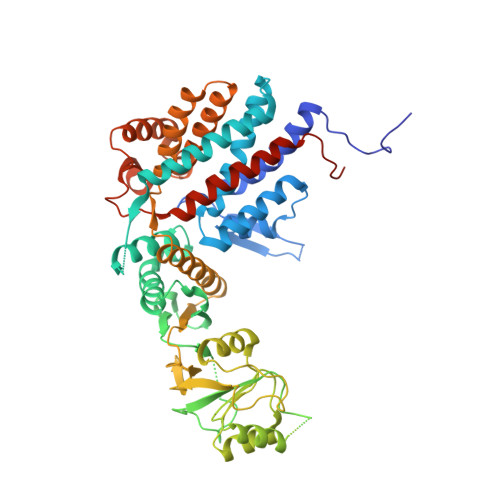
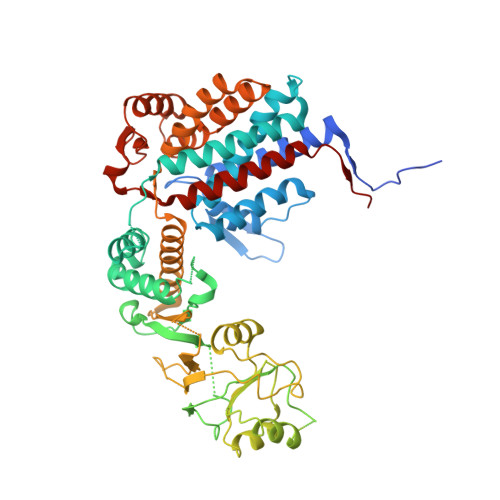
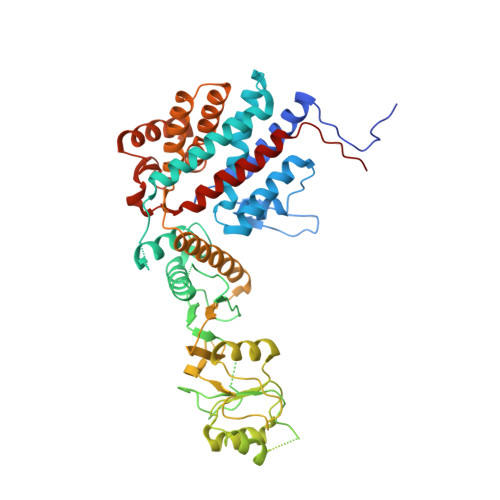
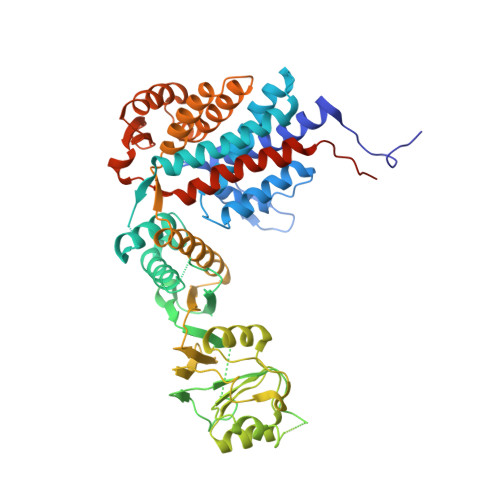
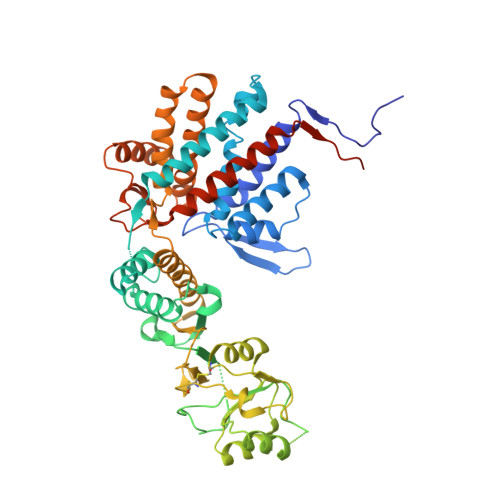
The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Kalisman, N., Schroder, G.F., Levitt, M.(2013) Structure 21: 540
- PubMed: 23478063
- DOI: https://doi.org/10.1016/j.str.2013.01.017
- Primary Citation of Related Structures:
4B2T, 4V8R - PubMed Abstract:
In eukaryotes, CCT is essential for the correct and efficient folding of many cytosolic proteins, most notably actin and tubulin. Structural studies of CCT have been hindered by the failure of standard crystallographic analysis to resolve its eight different subunit types at low resolutions. Here, we exhaustively assess the R value fit of all possible CCT models to available crystallographic data of the closed and open forms with resolutions of 3.8 Å and 5.5 Å, respectively. This unbiased analysis finds the native subunit arrangements with overwhelming significance. The resulting structures provide independent crystallographic proof of the subunit arrangement of CCT and map major asymmetrical features of the particle onto specific subunits. The actin and tubulin substrates both bind around subunit CCT6, which shows other structural anomalies. CCT is thus clearly partitioned, both functionally and evolutionary, into a substrate-binding side that is opposite to the ATP-hydrolyzing side.
- Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: