Akt-Mediated Phosphorylation Increases the Binding Affinity of Htert for Importin Alpha to Promote Nuclear Translocation.
Jeong, S.A., Kim, K., Lee, J.H., Cha, J.S., Khadka, P., Cho, H., Chung, I.K.(2015) J Cell Sci 128: 2287
- PubMed: 25999477
- DOI: https://doi.org/10.1242/jcs.166132
- Primary Citation of Related Structures:
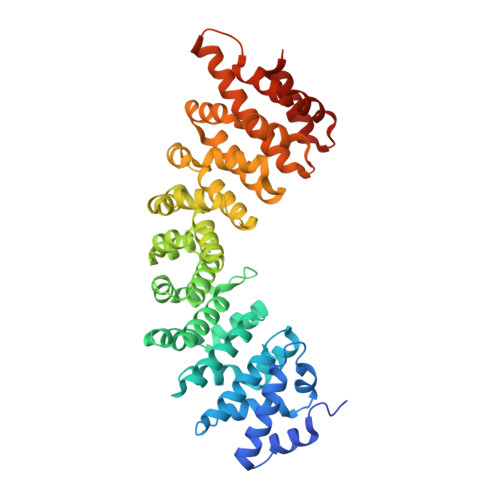
4B18 - PubMed Abstract:
Telomeres are essential for chromosome integrity and protection, and their maintenance requires the ribonucleoprotein enzyme telomerase. Previously, we have shown that human telomerase reverse transcriptase (hTERT) contains a bipartite nuclear localization signal (NLS; residues 222-240) that is responsible for nuclear import, and that Akt-mediated phosphorylation of residue S227 is important for efficient nuclear import of hTERT. Here, we show that hTERT binds to importin-α proteins through the bipartite NLS and that this heterodimer then forms a complex with importin-β proteins to interact with the nuclear pore complex. Depletion of individual importin-α proteins results in a failure of hTERT nuclear import, and the resulting cytoplasmic hTERT is degraded by ubiquitin-dependent proteolysis. Crystallographic analysis reveals that the bipartite NLS interacts with both the major and minor sites of importin-α proteins. We also show that Akt-mediated phosphorylation of S227 increases the binding affinity for importin-α proteins and promotes nuclear import of hTERT, thereby resulting in increased telomerase activity. These data provide details of a binding mechanism that enables hTERT to interact with the nuclear import receptors and of the control of the dynamic nuclear transport of hTERT through phosphorylation.
- Department of Integrated Omics for Biomedical Science, Yonsei University, Seoul 120-749, Korea.
Organizational Affiliation: