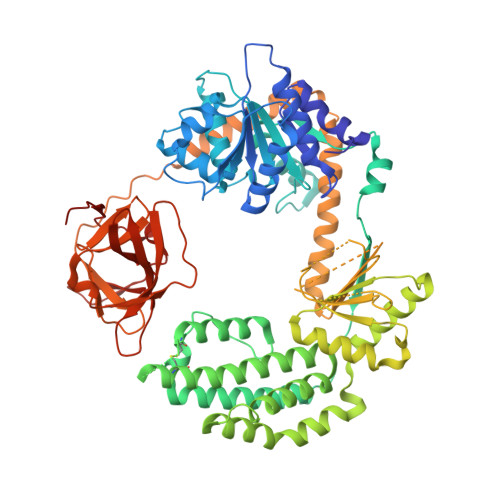
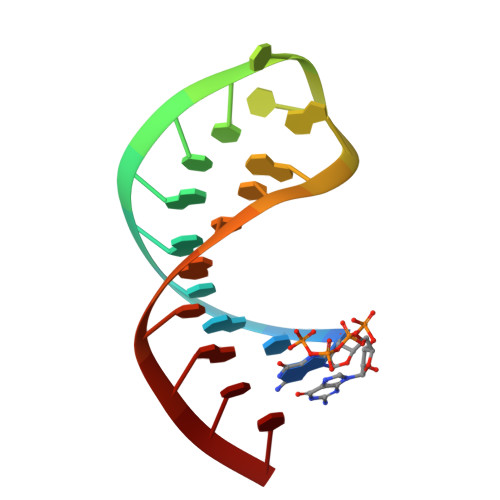
Visualizing the Determinants of Viral RNA Recognition by Innate Immune Sensor Rig-I.
Luo, D., Kohlway, A., Vela, A., Pyle, A.M.(2012) Structure 20: 1983
- PubMed: 23022350
- DOI: https://doi.org/10.1016/j.str.2012.08.029
- Primary Citation of Related Structures:
4AY2 - PubMed Abstract:
Retinoic acid inducible gene-I (RIG-I) is a key intracellular immune receptor for pathogenic RNAs, particularly from RNA viruses. Here, we report the crystal structure of human RIG-I bound to a 5' triphosphorylated RNA hairpin and ADP nucleotide at 2.8 Å resolution. The RNA ligand contains all structural features that are essential for optimal recognition by RIG-I, as it mimics the panhandle-like signatures within the genome of negative-stranded RNA viruses. RIG-I adopts an intermediate, semiclosed conformation in this product state of ATP hydrolysis. The structure of this complex allows us to visualize the first steps in RIG-I recognition and activation upon viral infection.
- Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT 06520, USA.
Organizational Affiliation: