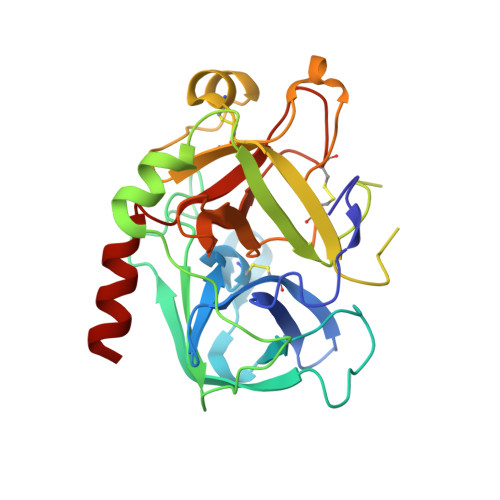
Design and Synthesis of Potent and Highly Selective Thrombin Inhibitors.
Hilpert, K., Ackermann, J., Banner, D.W., Gast, A., Gubernator, K., Hadvary, P., Labler, L., Muller, K., Schmid, G., Tschopp, T.B.(1994) J Med Chem 37: 3889
- PubMed: 7966150
- DOI: https://doi.org/10.1021/jm00049a008
- Primary Citation of Related Structures:
4AX9, 4AYV, 4AYY, 4AZ2 - PubMed Abstract:
Thrombin, a serine protease, plays a central role in the initiation and propagation of thrombotic events. An extensive search for new thrombin inhibitors was performed, using an unconventional approach. Screening of small basic molecules for binding in the recognition pocket of thrombin led to the discovery of (aminoiminomethyl)piperidine (amidinopiperidine) as a weak, but intrinsically selective, thrombin inhibitor. Elaboration of this molecule provided compounds which inhibit thrombin with Ki's in the range of 20-50 nM and with selectivities of 1000-4000 against trypsin. These inhibitor compounds show a new and unexpected binding mode to thrombin. Modification of the central building block and then of one of the hydrophobic substituents led to the discovery of a new family of thrombin inhibitors which has reverted to the former binding mode to thrombin. This last class of compounds shows inhibitory activities in the picomolar range, low toxicity, and a short plasma half life which favors its use for an intravenous application. From this series of thrombin inhibitors, 19f(Ro 46-6240) was selected for clinical development as an antithrombotic agent for intravenous administration.
- Pharma Division, F. Hoffmann-La Roche Ltd., Basel, Switzerland.
Organizational Affiliation: