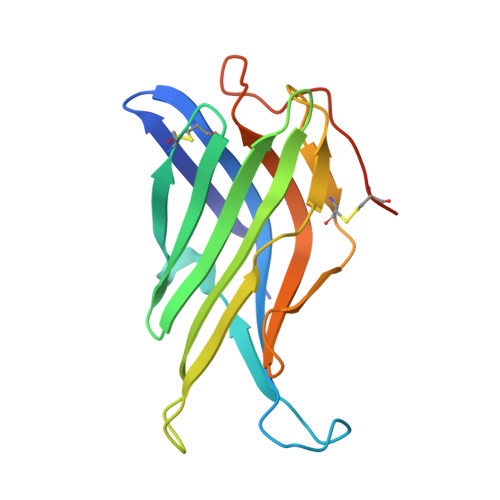
Crystal Structure of the Conserved Domain of the Dc Lysosomal Associated Membrane Protein: Implications for the Lysosomal Glycocalyx.
Wilke, S., Krausze, J., Bussow, K.(2012) BMC Biol 10: 62
- PubMed: 22809326
- DOI: https://doi.org/10.1186/1741-7007-10-62
- Primary Citation of Related Structures:
4AKM - PubMed Abstract:
The family of lysosome-associated membrane proteins (LAMP) comprises the multifunctional, ubiquitous LAMP-1 and LAMP-2, and the cell type-specific proteins DC-LAMP (LAMP-3), BAD-LAMP (UNC-46, C20orf103) and macrosialin (CD68). LAMPs have been implicated in a multitude of cellular processes, including phagocytosis, autophagy, lipid transport and aging. LAMP-2 isoform A acts as a receptor in chaperone-mediated autophagy. LAMP-2 deficiency causes the fatal Danon disease. The abundant proteins LAMP-1 and LAMP-2 are major constituents of the glycoconjugate coat present on the inside of the lysosomal membrane, the 'lysosomal glycocalyx'. The LAMP family is characterized by a conserved domain of 150 to 200 amino acids with two disulfide bonds. The crystal structure of the conserved domain of human DC-LAMP was solved. It is the first high-resolution structure of a heavily glycosylated lysosomal membrane protein. The structure represents a novel β-prism fold formed by two β-sheets bent by β-bulges and connected by a disulfide bond. Flexible loops and a hydrophobic pocket represent possible sites of molecular interaction. Computational models of the glycosylated luminal regions of LAMP-1 and LAMP-2 indicate that the proteins adopt a compact conformation in close proximity to the lysosomal membrane. The models correspond to the thickness of the lysosomal glycoprotein coat of only 5 to 12 nm, according to electron microscopy. The conserved luminal domain of lysosome-associated membrane proteins forms a previously unknown β-prism fold. Insights into the structure of the lysosomal glycoprotein coat were obtained by computational models of the LAMP-1 and LAMP-2 luminal regions.
- Department of Molecular Structural Biology, Helmholtz Centre for Infection Research, Inhoffenstr, 7, 38124 Braunschweig, Germany.
Organizational Affiliation: