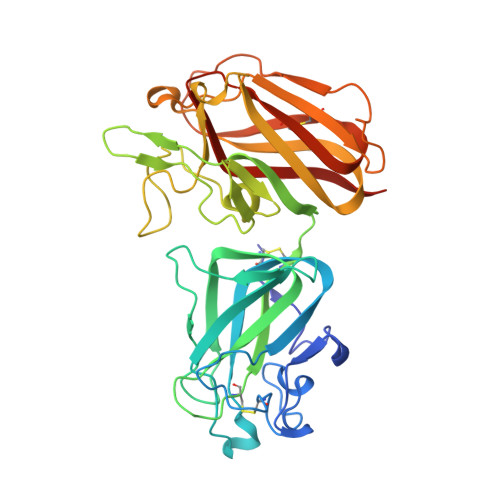
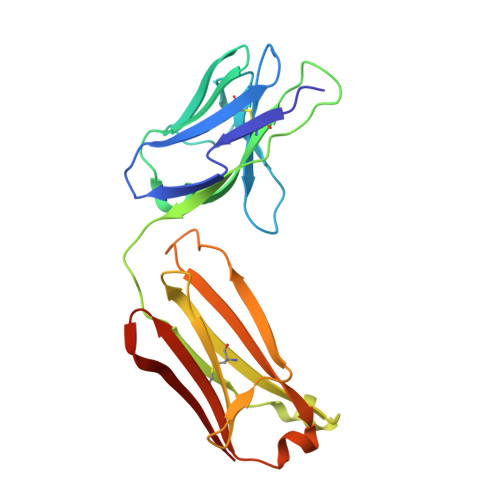
Structure of the Discoidin Domain Receptor 1 Extracellular Region Bound to an Inhibitory Fab Fragment Reveals Features Important for Signaling.
Carafoli, F., Mayer, M.C., Shiraishi, K., Pecheva, M.A., Chan, L.Y., Nan, R., Leitinger, B., Hohenester, E.(2012) Structure 20: 688
- PubMed: 22483115
- DOI: https://doi.org/10.1016/j.str.2012.02.011
- Primary Citation of Related Structures:
4AG4 - PubMed Abstract:
The discoidin domain receptors, DDR1 and DDR2, are constitutively dimeric receptor tyrosine kinases that are activated by triple-helical collagen. Aberrant DDR signaling contributes to several human pathologies, including many cancers. We have generated monoclonal antibodies (mAbs) that inhibit DDR1 signaling without interfering with collagen binding. The crystal structure of the monomeric DDR1 extracellular region bound to the Fab fragment of mAb 3E3 reveals that the collagen-binding discoidin (DS) domain is tightly associated with the following DS-like domain, which contains the epitopes of all mAbs. A conserved surface patch in the DS domain outside the collagen-binding site is shown to be required for signaling. Thus, the active conformation of the DDR1 dimer involves collagen-induced contacts between the DS domains, in addition to the previously identified association of transmembrane helices. The mAbs likely inhibit signaling by sterically blocking the extracellular association of DDR1 subunits.
- Department of Life Sciences, Imperial College London, London SW7 2AZ, UK.
Organizational Affiliation: