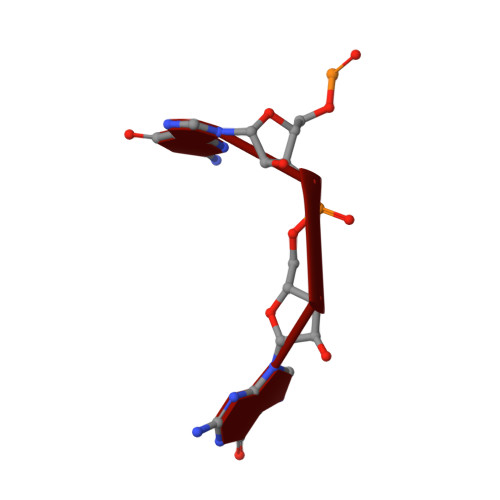
Crystal Structure of an Eal Domain in Complex with Reaction Product 5'-Pgpg
Robert-Paganin, J., Nonin-Lecomte, S., Rety, S.(2012) PLoS One 7: 52424
- PubMed: 23285035
- DOI: https://doi.org/10.1371/journal.pone.0052424
- Primary Citation of Related Structures:
4AFY, 4AG0 - PubMed Abstract:
FimX is a large multidomain protein containing an EAL domain and involved in twitching motility in Pseudomonas aeruginosa. We present here two crystallographic structures of the EAL domain of FimX (residues 438-686): one of the apo form and the other of a complex with 5'-pGpG, the reaction product of the hydrolysis of c-di-GMP. In both crystal forms, the EAL domains form a dimer delimiting a large cavity encompassing the catalytic pockets. The ligand is trapped in this cavity by its sugar phosphate moiety. We confirmed by NMR that the guanine bases are not involved in the interaction in solution. We solved here the first structure of an EAL domain bound to the reaction product 5'-pGpG. Though isolated FimX EAL domain has a very low catalytic activity, which would not be significant compared to other catalytic EAL domains, the structure with the product of the reaction can provides some hints in the mechanism of hydrolysis of the c-di-GMP by EAL domains.
- Laboratoire de Cristallographie et RMN biologiques, UMR 8015-Cente National de la Recherche Scientifique, Paris, France.
Organizational Affiliation: