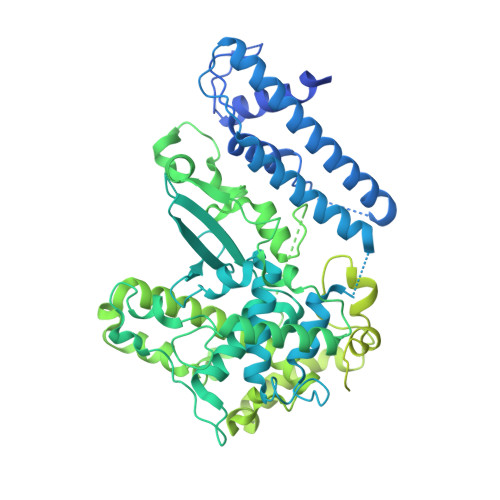
The Structure of the Yeast Kinetochore Ndc10 DNA-Binding Domain Reveals an Unexpected Evolutionary Relationship to Tyrosine Recombinases.
Perriches, T., Singleton, M.R.(2012) J Biological Chem 287: 5173
- PubMed: 22215672
- DOI: https://doi.org/10.1074/jbc.C111.318501
- Primary Citation of Related Structures:
4ACO - PubMed Abstract:
We have solved the x-ray structure of the N-terminal half of the yeast kinetochore protein Ndc10 at 1.9 Å resolution. This essential protein is a key constituent of the budding yeast centromere and is essential for the recruitment of the centromeric nucleosome and establishment of the kinetochore. The fold of the protein shows unexpected similarities to the tyrosine recombinase/λ-integrase family of proteins, most notably Cre, with some variation in the relative position of the subdomains. This finding offers new insights into kinetochore evolution and the adaptation of a well studied protein fold to a novel role. By comparison with tyrosine recombinases and mutagenesis studies, we have been able to define some of the key DNA-binding motifs.
- Macromolecular Structure and Function Laboratory, Cancer Research UK London Research Institute, 44 Lincoln's Inn Fields, London WC2A 3LY, United Kingdom.
Organizational Affiliation: