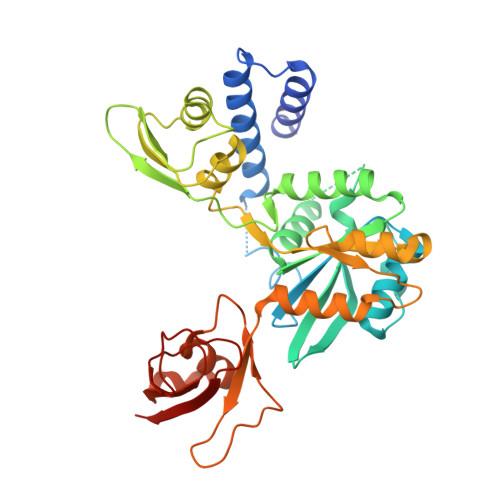
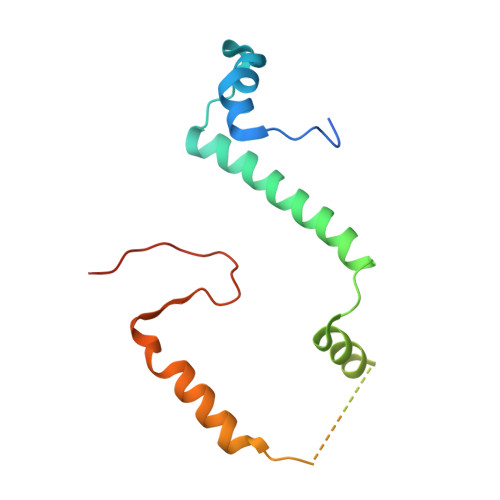
Rbg1-Tma46 Dimer Structure Reveals New Functional Domains and Their Role in Polysome Recruitment.
Francis, S.M., Gas, M., Daugeron, M., Bravo, J., Seraphin, B.(2012) Nucleic Acids Res 40: 11100
- PubMed: 23002146
- DOI: https://doi.org/10.1093/nar/gks867
- Primary Citation of Related Structures:
4A9A - PubMed Abstract:
Developmentally Regulated GTP-binding (DRG) proteins are highly conserved GTPases that associate with DRG Family Regulatory Proteins (DFRP). The resulting complexes have recently been shown to participate in eukaryotic translation. The structure of the Rbg1 GTPase, a yeast DRG protein, in complex with the C-terminal region of its DFRP partner, Tma46, was solved by X-ray diffraction. These data reveal that DRG proteins are multimodular factors with three additional domains, helix-turn-helix (HTH), S5D2L and TGS, packing against the GTPase platform. Surprisingly, the S5D2L domain is inserted in the middle of the GTPase sequence. In contrast, the region of Tma46 interacting with Rbg1 adopts an extended conformation typical of intrinsically unstructured proteins and contacts the GTPase and TGS domains. Functional analyses demonstrate that the various domains of Rbg1, as well as Tma46, modulate the GTPase activity of Rbg1 and contribute to the function of these proteins in vivo. Dissecting the role of the different domains revealed that the Rbg1 TGS domain is essential for the recruitment of this factor in polysomes, supporting further the implication of these conserved factors in translation.
- Instituto de Biomedicina de Valencia (IBV-CSIC), Calle Jaime Roig, 11, Valencia E-46010, Spain.
Organizational Affiliation: