Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
Wright, S., Poranen, M.M., Bamford, D.H., Stuart, D.I., Grimes, J.M.(2012) J Virol 86: 2837
- PubMed: 22205747
- DOI: https://doi.org/10.1128/JVI.05168-11
- Primary Citation of Related Structures:
4A8F, 4A8K, 4A8M, 4A8O, 4A8Q, 4A8S, 4A8W, 4A8Y - PubMed Abstract:
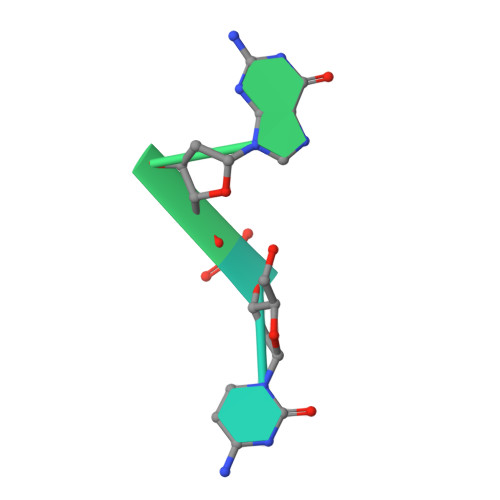
RNA-dependent RNA polymerases (RdRps) are key to the replication of RNA viruses. A common divalent cation binding site, distinct from the positions of catalytic ions, has been identified in many viral RdRps. We have applied biochemical, biophysical, and structural approaches to show how the RdRp from bacteriophage ϕ6 uses the bound noncatalytic Mn(2+) to facilitate the displacement of the C-terminal domain during the transition from initiation to elongation. We find that this displacement releases the noncatalytic Mn(2+), which must be replaced for elongation to occur. By inserting a dysfunctional Mg(2+) at this site, we captured two nucleoside triphosphates within the active site in the absence of Watson-Crick base pairing with template and mapped movements of divalent cations during preinitiation. These structures refine the pathway from preinitiation through initiation to elongation for the RNA-dependent RNA polymerization reaction, explain the role of the noncatalytic divalent cation in 6 RdRp, and pinpoint the previously unresolved Mn(2+)-dependent step in replication.
- Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: