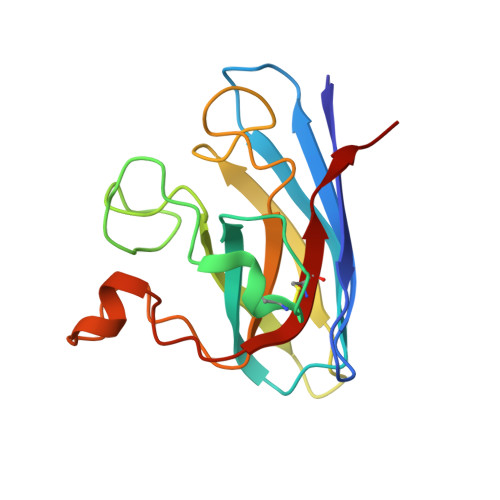
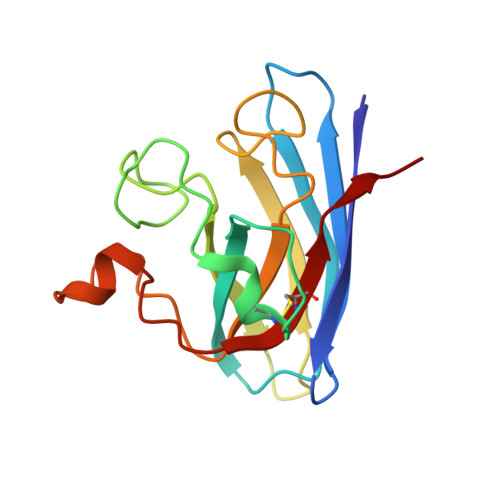
X-ray crystallography and computational docking for the detection and development of protein-ligand interactions.
Kershaw, N.M., Wright, G.S., Sharma, R., Antonyuk, S.V., Strange, R.W., Berry, N.G., O'Neill, P.M., Hasnain, S.S.(2013) Curr Med Chem 20: 569-575
- PubMed: 23278398
- DOI: https://doi.org/10.2174/0929867311320040008
- Primary Citation of Related Structures:
4A7G, 4A7Q - PubMed Abstract:
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disorder characterised by the selective dysfunction and death of the upper and lower motor neurons. Median survival rates are between 3 and 5 years after diagnosis. Mutations in the gene encoding Cu/Zn superoxide dismutase (SOD1) have been linked to a subset of familial forms of ALS (fALS). Herein, we describe a fragment- based drug discovery (FBDD) approach for the investigation of small molecule binding sites in SOD1. X-ray crystallography has been used as the primary screening method and has been shown to directly detect protein-ligand interactions which cannot be unambiguously identified using other biophysical methods. The structural requirements for effective binding at Trp32 are detailed for a series of quinazoline-containing compounds. The investigation of an additional site that binds a range of catecholamines and the use of computational modelling to assist fragment evolution is discussed. This study also highlights the importance of ligand solubility for successful Xray crystallographic campaigns in lead compound design.
- Molecular Biophysics Group, Institute of Integrative Biology, Faculty of Health and Life Sciences, University of Liverpool, Liverpool, UK. nkershaw@liverpool.ac.uk
Organizational Affiliation: