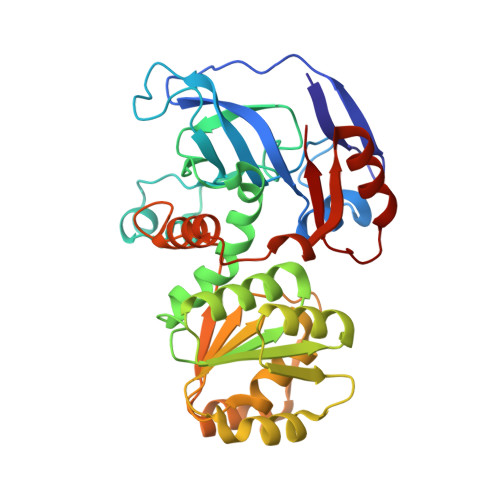
The Crystal Structure of Galactitol-1-Phosphate 5-Dehydrogenase from Escherichia Coli K12 Provides Insights Into its Anomalous Behavior on Imac Processes
Esteban-Torres, M., Alvarez, Y., Acebron, I., De Las Rivas, B., Munoz, R., Kohring, G.-W., Roa, A.M., Sobrino, M., Mancheno, J.M.(2012) FEBS Lett 586: 3127
- PubMed: 22979983
- DOI: https://doi.org/10.1016/j.febslet.2012.07.073
- Primary Citation of Related Structures:
4A2C - PubMed Abstract:
Endogenous galactitol-1-phosphate 5-dehydrogenase (GPDH) (EC 1.1.1.251) from Escherichia coli spontaneously interacts with Ni(2+)-NTA matrices becoming a potential contaminant for recombinant, target His-tagged proteins. Purified recombinant, untagged GPDH (rGPDH) converted galactitol into tagatose, and d-tagatose-6-phosphate into galactitol-1-phosphate, in a Zn(2+)- and NAD(H)-dependent manner and readily crystallized what has permitted to solve its crystal structure. In contrast, N-terminally His-tagged GPDH was marginally stable and readily aggregated. The structure of rGPDH revealed metal-binding sites characteristic from the medium-chain dehydrogenase/reductase protein superfamily which may explain its ability to interact with immobilized metals. The structure also provides clues on the harmful effects of the N-terminal His-tag.
- Laboratorio de Biotecnología Bacteriana, Instituto Ciencia y Tecnología de Alimentos y Nutrición (ICTAN), CSIC, Madrid, Spain.
Organizational Affiliation: