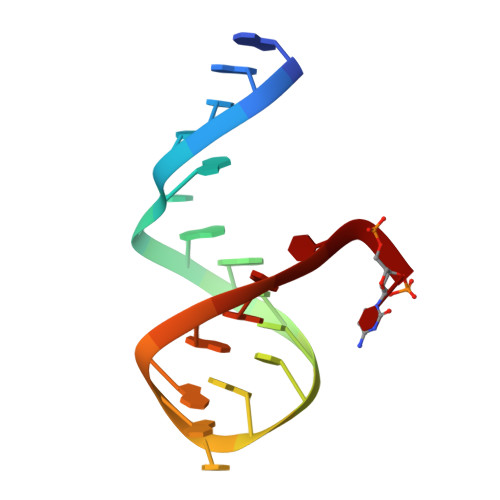
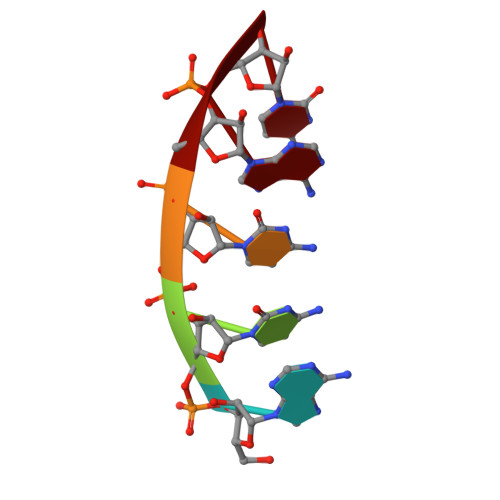
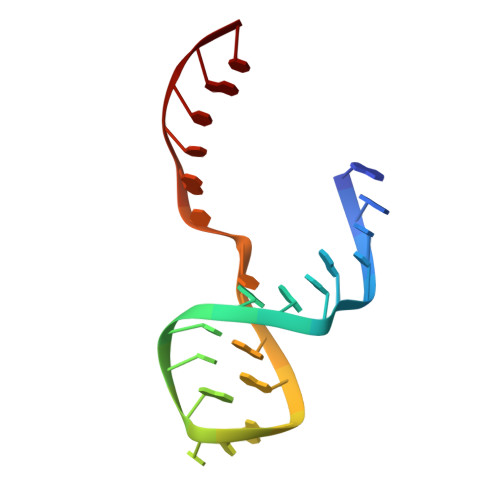
Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction.
Murray, J.B., Szoke, H., Szoke, A., Scott, W.G.(2000) Mol Cell 5: 279-287
- PubMed: 10882069
- DOI: https://doi.org/10.1016/s1097-2765(00)80423-2
- Primary Citation of Related Structures:
488D - PubMed Abstract:
We have determined the crystal structure of the enzyme-product complex of the hammerhead ribozyme by using a reinforced crystal lattice to trap the complex prior to dissociation and by employing X-ray holographic image reconstruction, a real-space electron density imaging and refinement procedure. Subsequent to catalysis, the cleavage site residue (C-17), together with its 2',3'-cyclic phosphate, adopts a conformation close to and approximately perpendicular to the Watson-Crick base-pairing faces of two highly conserved purines in the ribozyme's catalytic pocket (G-5 and A-6). We observe several interactions with functional groups on these residues that have been identified as critical for ribozyme activity by biochemical analyses but whose role has defied explanation in terms of previous structural analyses. These interactions may therefore be relevant to the hammerhead ribozyme reaction mechanism.
- The Center for the Molecular Biology of RNA and Department of Chemistry and Biochemistry, University of California, Santa Cruz 95064, USA.
Organizational Affiliation: