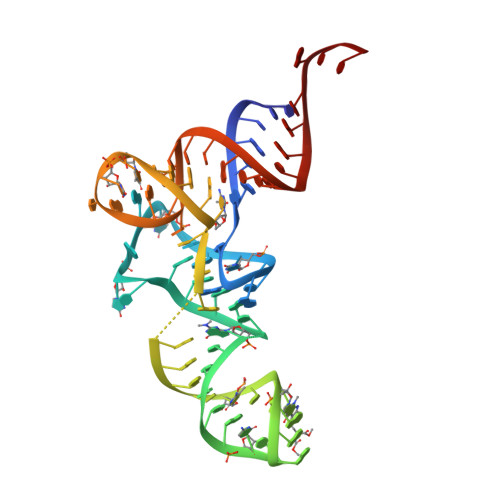
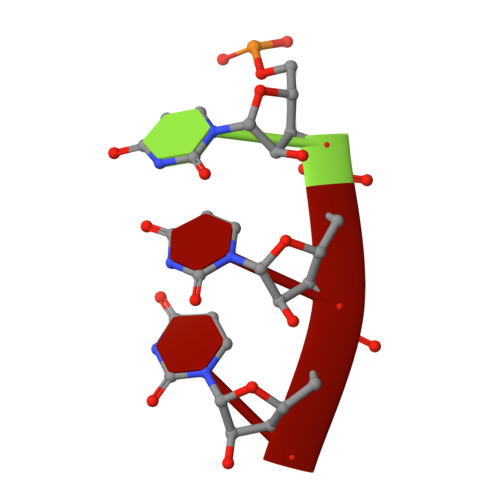
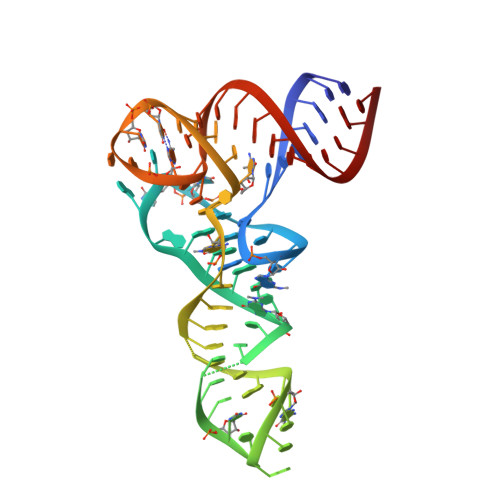
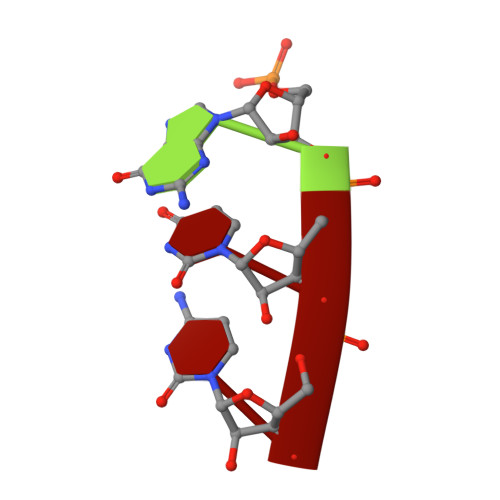
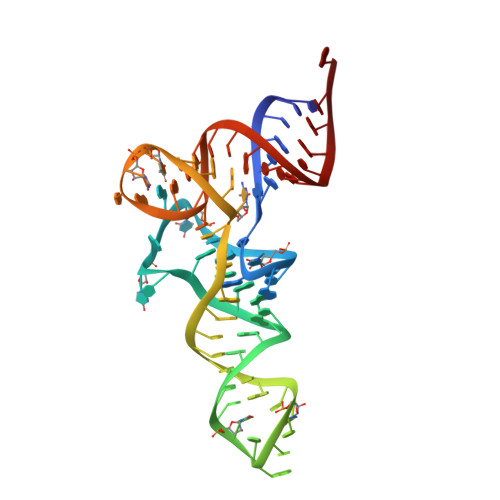
X-ray crystal structures of 70S ribosome functional complexes.
Cate, J.H., Yusupov, M.M., Yusupova, G.Z., Earnest, T.N., Noller, H.F.(1999) Science 285: 2095-2104
- PubMed: 10497122
- DOI: https://doi.org/10.1126/science.285.5436.2095
- Primary Citation of Related Structures:
486D - PubMed Abstract:
Structures of 70S ribosome complexes containing messenger RNA and transfer RNA (tRNA), or tRNA analogs, have been solved by x-ray crystallography at up to 7.8 angstrom resolution. Many details of the interactions between tRNA and the ribosome, and of the packing arrangements of ribosomal RNA (rRNA) helices in and between the ribosomal subunits, can be seen. Numerous contacts are made between the 30S subunit and the P-tRNA anticodon stem-loop; in contrast, the anticodon region of A-tRNA is much more exposed. A complex network of molecular interactions suggestive of a functional relay is centered around the long penultimate stem of 16S rRNA at the subunit interface, including interactions involving the "switch" helix and decoding site of 16S rRNA, and RNA bridges from the 50S subunit.
- Center for Molecular Biology of RNA, Sinsheimer Laboratories, University of California, Santa Cruz, CA 95064, USA. cate@wi.mit.edu
Organizational Affiliation: