Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea.
Liu, Y., Nakamura, A., Nakazawa, Y., Asano, N., Ford, K.A., Hohn, M.J., Tanaka, I., Yao, M., Soll, D.(2014) Proc Natl Acad Sci U S A 111: 10520-10525
- PubMed: 25002468
- DOI: https://doi.org/10.1073/pnas.1411267111
- Primary Citation of Related Structures:
3WKR, 3WKS - PubMed Abstract:
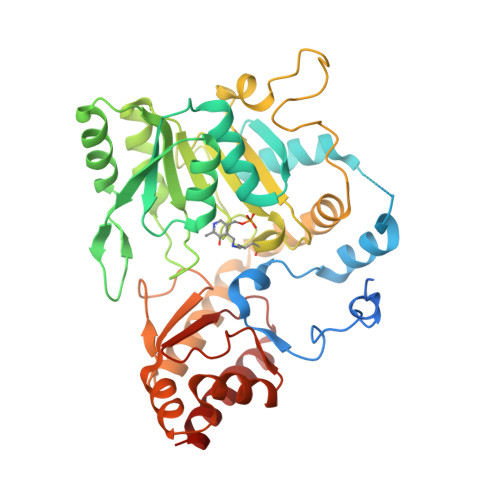
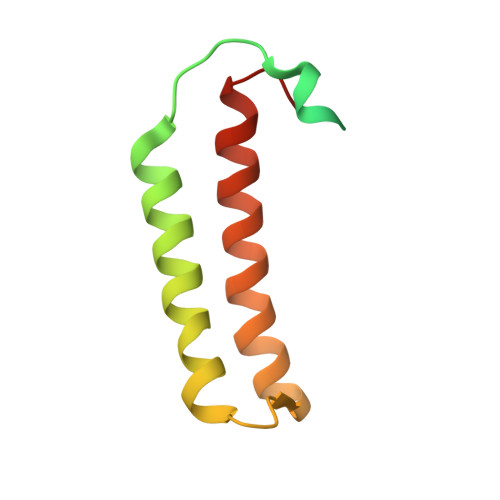
Methanogenic archaea lack cysteinyl-tRNA synthetase; they synthesize Cys-tRNA and cysteine in a tRNA-dependent manner. Two enzymes are required: Phosphoseryl-tRNA synthetase (SepRS) forms phosphoseryl-tRNA(Cys) (Sep-tRNA(Cys)), which is converted to Cys-tRNA(Cys) by Sep-tRNA:Cys-tRNA synthase (SepCysS). This represents the ancestral pathway of Cys biosynthesis and coding in archaea. Here we report a translation factor, SepCysE, essential for methanococcal Cys biosynthesis; its deletion in Methanococcus maripaludis causes Cys auxotrophy. SepCysE acts as a scaffold for SepRS and SepCysS to form a stable high-affinity complex for tRNA(Cys) causing a 14-fold increase in the initial rate of Cys-tRNA(Cys) formation. Based on our crystal structure (2.8-Å resolution) of a SepCysS⋅SepCysE complex, a SepRS⋅SepCysE⋅SepCysS structure model suggests that this ternary complex enables substrate channeling of Sep-tRNA(Cys). A phylogenetic analysis suggests coevolution of SepCysE with SepRS and SepCysS in the last universal common ancestral state. Our findings suggest that the tRNA-dependent Cys biosynthesis proceeds in a multienzyme complex without release of the intermediate and this mechanism may have facilitated the addition of Cys to the genetic code.
- Departments of Molecular Biophysics and Biochemistry and.
Organizational Affiliation: