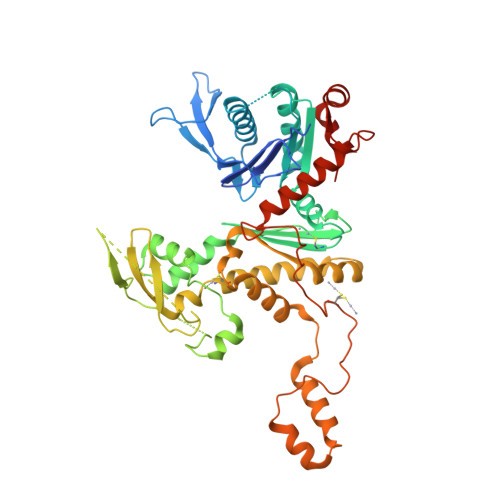
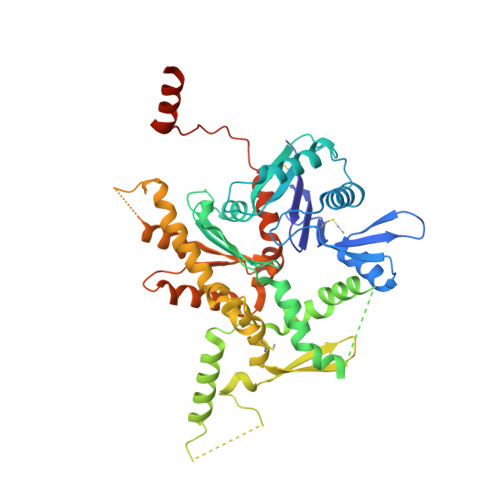
Structure of the full-length yeast Arp7-Arp9 heterodimer.
Lobsiger, J., Hunziker, Y., Richmond, T.J.(2014) Acta Crystallogr D Biol Crystallogr 70: 310-316
- PubMed: 24531465
- DOI: https://doi.org/10.1107/S1399004713027417
- Primary Citation of Related Structures:
3WEE - PubMed Abstract:
The nuclear actin-related proteins Arp7 and Arp9 are components of the yeast SWI/SNF and RSC chromatin-remodelling complexes. The 3.1 Å resolution crystal structure reported here shows that the full-length Arp7 and Arp9 proteins exist as a dimer without a requirement for additional polypeptides. Of the 11 actin-related proteins, Arp7 and Arp9 are the only two directly demonstrated to form a dimer within this family. The Arp7-Arp9 heterodimer is unlikely to form an actin-like filament based on modelling using the structure. The Arp7-Arp9 structure reveals that its dimerization interface is not altered when bound in a complex with the SWI/SNF Snf2 HSA domain and the regulatory protein Rtt102.
- Institute of Molecular Biology and Biophysics, Department of Biology, ETH Zürich, Schafmattstrasse 20, ETH-Hönggerberg, CH-8093 Zürich, Switzerland.
Organizational Affiliation: