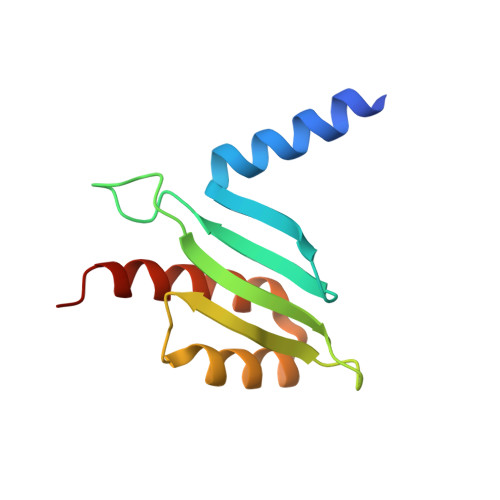
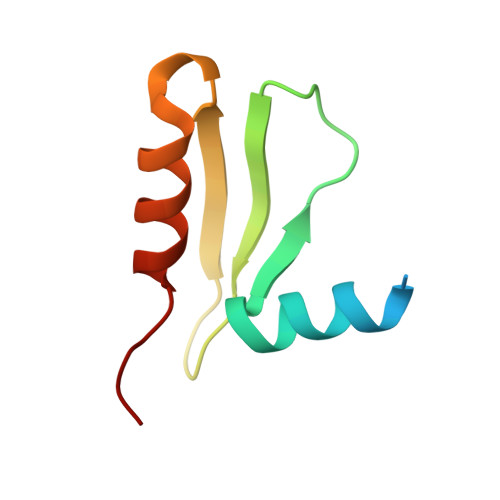
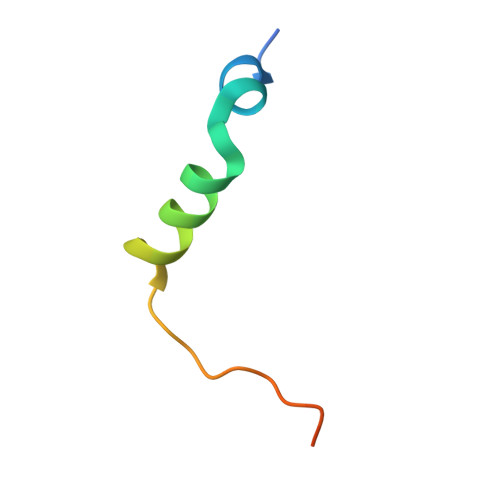
CENP-T provides a structural platform for outer kinetochore assembly
Nishino, T., Rago, F., Hori, T., Tomii, K., Cheeseman, I.M., Fukagawa, T.(2013) EMBO J 32: 424-436
- PubMed: 23334297
- DOI: https://doi.org/10.1038/emboj.2012.348
- Primary Citation of Related Structures:
3VZ9, 3VZA - PubMed Abstract:
The kinetochore forms a dynamic interface with microtubules from the mitotic spindle during mitosis. The Ndc80 complex acts as the key microtubule-binding complex at kinetochores. However, it is unclear how the Ndc80 complex associates with the inner kinetochore proteins that assemble upon centromeric chromatin. Here, based on a high-resolution structural analysis, we demonstrate that the N-terminal region of vertebrate CENP-T interacts with the 'RWD' domain in the Spc24/25 portion of the Ndc80 complex. Phosphorylation of CENP-T strengthens a cryptic hydrophobic interaction between CENP-T and Spc25 resulting in a phospho-regulated interaction that occurs without direct recognition of the phosphorylated residue. The Ndc80 complex interacts with both CENP-T and the Mis12 complex, but we find that these interactions are mutually exclusive, supporting a model in which two distinct pathways target the Ndc80 complex to kinetochores. Our results provide a model for how the multiple protein complexes at kinetochores associate in a phospho-regulated manner.
- Department of Molecular Genetics, National Institute of Genetics and The Graduate University for Advanced Studies SOKENDAI, Shizuoka 411-8540, Japan.
Organizational Affiliation: