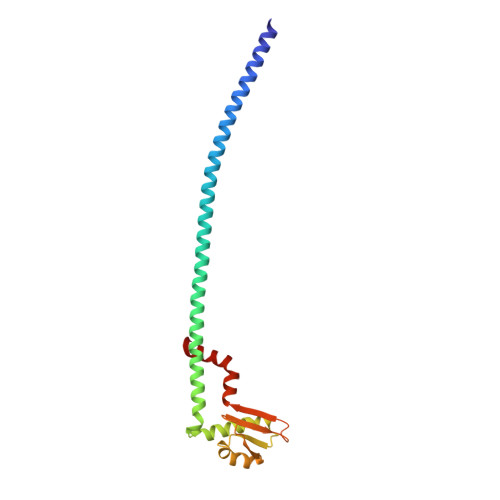
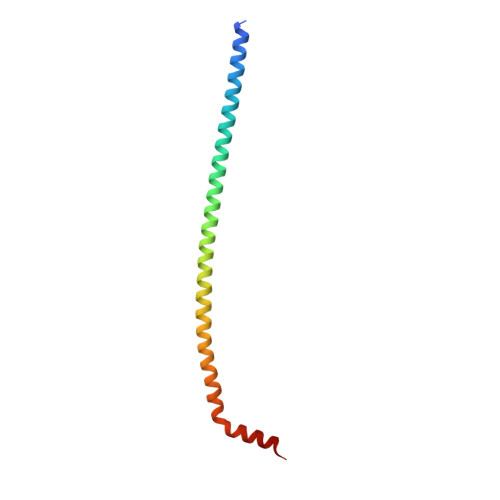
The dynamic stator stalk of rotary ATPases
Stewart, A.G., Lee, L.K., Donohoe, M., Chaston, J.J., Stock, D.(2012) Nat Commun 3: 687-687
- PubMed: 22353718
- DOI: https://doi.org/10.1038/ncomms1693
- Primary Citation of Related Structures:
3V6I - PubMed Abstract:
Rotary ATPases couple ATP hydrolysis/synthesis with proton translocation across biological membranes and so are central components of the biological energy conversion machinery. Their peripheral stalks are essential components that counteract torque generated by rotation of the central stalk during ATP synthesis or hydrolysis. Here we present a 2.25-Å resolution crystal structure of the peripheral stalk from Thermus thermophilus A-type ATPase/synthase. We identify bending and twisting motions inherent within the structure that accommodate and complement a radial wobbling of the ATPase headgroup as it progresses through its catalytic cycles, while still retaining azimuthal stiffness necessary to counteract rotation of the central stalk. The conformational freedom of the peripheral stalk is dictated by its unusual right-handed coiled-coil architecture, which is in principle conserved across all rotary ATPases. In context of the intact enzyme, the dynamics of the peripheral stalks provides a potential mechanism for cooperativity between distant parts of rotary ATPases.
- Structural and Computational Biology Division, The Victor Chang Cardiac Research Institute, Darlinghurst, Australia.
Organizational Affiliation: