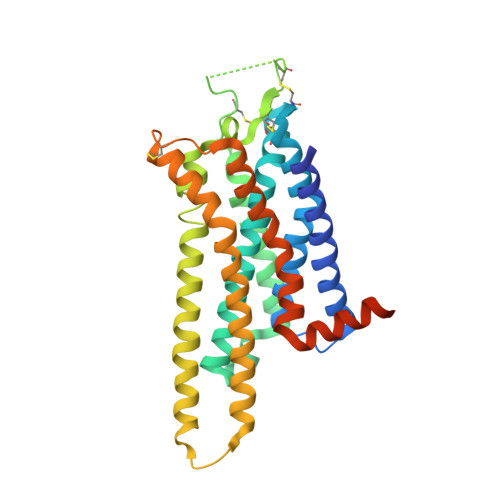
Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design
Congreve, M., Andrews, S.P., Dore, A.S., Hollenstein, K., Hurrell, E., Langmead, C.J., Mason, J.S., Ng, I.W., Tehan, B., Zhukov, A., Weir, M., Marshall, F.H.(2012) J Med Chem 55: 1898-1903
- PubMed: 22220592
- DOI: https://doi.org/10.1021/jm201376w
- Primary Citation of Related Structures:
3UZA, 3UZC - PubMed Abstract:
Potent, ligand efficient, selective, and orally efficacious 1,2,4-triazine derivatives have been identified using structure based drug design approaches as antagonists of the adenosine A(2A) receptor. The X-ray crystal structures of compounds 4e and 4g bound to the GPCR illustrate that the molecules bind deeply inside the orthosteric binding cavity. In vivo pharmacokinetic and efficacy data for compound 4k are presented, demonstrating the potential of this series of compounds for the treatment of Parkinson's disease.
- Heptares Therapeutics Limited, BioPark, Broadwater Road, Welwyn Garden City, Hertfordshire AL7 3AX, UK. miles.congreve@heptares.com
Organizational Affiliation: