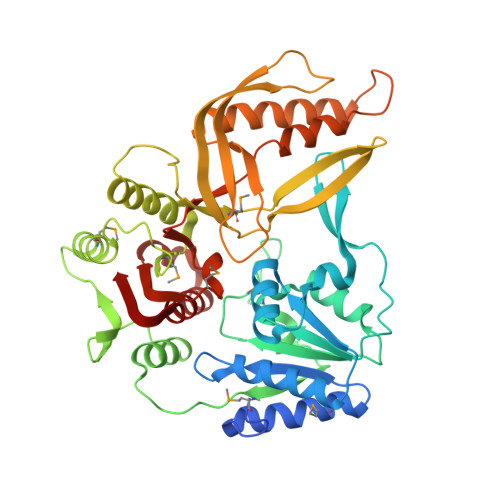
The T4 Phage SF1B Helicase Dda Is Structurally Optimized to Perform DNA Strand Separation.
He, X., Byrd, A.K., Yun, M.K., Pemble, C.W., Harrison, D., Yeruva, L., Dahl, C., Kreuzer, K.N., Raney, K.D., White, S.W.(2012) Structure 20: 1189-1200
- PubMed: 22658750
- DOI: https://doi.org/10.1016/j.str.2012.04.013
- Primary Citation of Related Structures:
3UPU - PubMed Abstract:
Helicases move on DNA via an ATP binding and hydrolysis mechanism coordinated by well-characterized helicase motifs. However, the translocation along single-stranded DNA (ssDNA) and the strand separation of double-stranded (dsDNA) may be loosely or tightly coupled. Dda is a phage T4 SF1B helicase with sequence homology to the Pif1 family of helicases that tightly couples translocation to strand separation. The crystal structure of the Dda-ssDNA binary complex reveals a domain referred to as the "pin" that was previously thought to remain static during strand separation. The pin contains a conserved phenylalanine that mediates a transient base-stacking interaction that is absolutely required for separation of dsDNA. The pin is secured at its tip by protein-protein interactions through an extended SH3 domain thereby creating a rigid strut. The conserved interface between the pin and the SH3 domain provides the mechanism for tight coupling of translocation to strand separation.
- Department of Structural Biology, St. Jude Children's Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105, USA.
Organizational Affiliation: