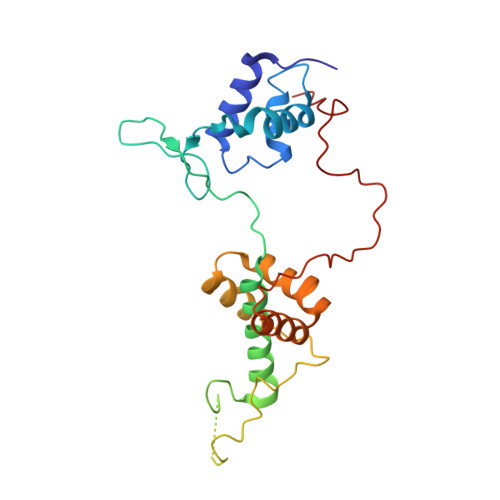
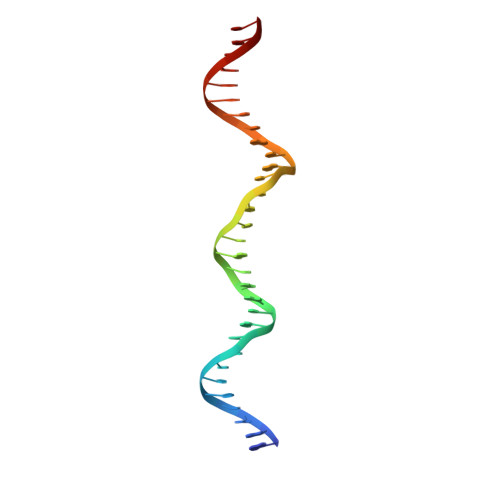
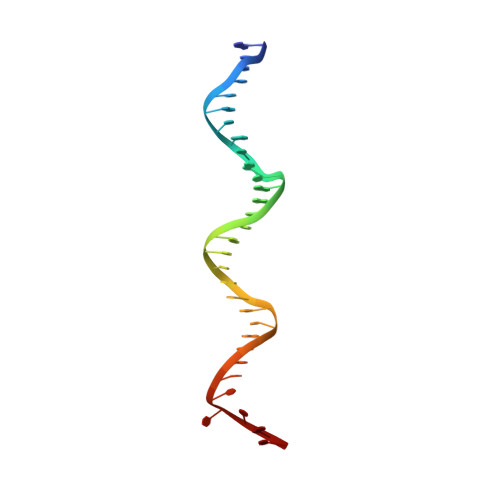
The orientation of the C-terminal domain of the Saccharomyces cerevisiae Rap1 protein is determined by its binding to DNA.
Matot, B., Le Bihan, Y.V., Lescasse, R., Perez, J., Miron, S., David, G., Castaing, B., Weber, P., Raynal, B., Zinn-Justin, S., Gasparini, S., Le Du, M.H.(2012) Nucleic Acids Res 40: 3197-3207
- PubMed: 22139930
- DOI: https://doi.org/10.1093/nar/gkr1166
- Primary Citation of Related Structures:
3UKG - PubMed Abstract:
Rap1 is an essential DNA-binding factor from the yeast Saccharomyces cerevisiae involved in transcription and telomere maintenance. Its binding to DNA targets Rap1 at particular loci, and may optimize its ability to form functional macromolecular assemblies. It is a modular protein, rich in large potentially unfolded regions, and comprising BRCT, Myb and RCT well-structured domains. Here, we present the architectures of Rap1 and a Rap1/DNA complex, built through a step-by-step integration of small angle X-ray scattering, X-ray crystallography and nuclear magnetic resonance data. Our results reveal Rap1 structural adjustment upon DNA binding that involves a specific orientation of the C-terminal (RCT) domain with regard to the DNA binding domain (DBD). Crystal structure of DBD in complex with a long DNA identifies an essential wrapping loop, which constrains the orientation of the RCT and affects Rap1 affinity to DNA. Based on our structural information, we propose a model for Rap1 assembly at telomere.
- Commissariat à l'Energie Atomique, Direction des Sciences du Vivant, Institut de Biologie et Technologie de Saclay, Laboratoire de Biologie Structurale et Radiobiologie, CNRS-URA2096, 91191 Gif-sur-Yvette, France.
Organizational Affiliation: