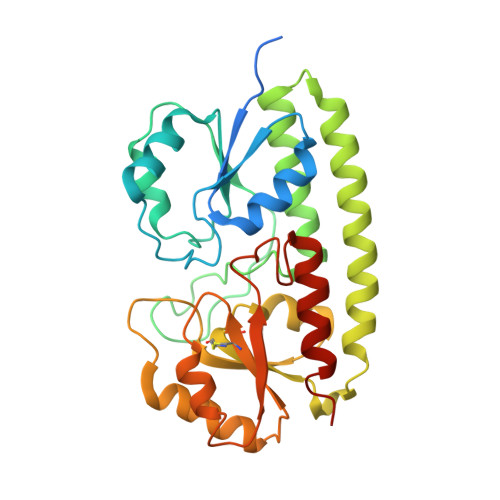
Arginine 116 stabilizes the entrance to the metal ion-binding site of the MntC protein.
Kanteev, M., Adir, N.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 237-242
- PubMed: 23519795
- DOI: https://doi.org/10.1107/S174430911300153X
- Primary Citation of Related Structures:
3UJP, 4IRM - PubMed Abstract:
The cyanobacterium Synechocystis sp. PCC 6803 imports Mn2+ ions via MntCAB, an ABC transport system that is expressed at submicromolar Mn2+ concentrations. The structures of the wild type (WT) and a site-directed mutant of the MntC solute-binding protein have been determined at 2.7 and 3.5 Å resolution, respectively. The WT structure is significantly improved over the previously determined structure (PDB entry 1xvl), showing improved Mn2+ binding site parameters, disulfide bonds in all three monomers and ions bound to the protein surface, revealing the role of Zn2+ ions in the crystallization liquor. The structure of MntC reveals that the active site is surrounded by neutral-to-positive electrostatic potential and is dominated by a network of polar interactions centred around Arg116. The mutation of this residue to alanine was shown to destabilize loops in the entrance to the metal-ion binding site and suggests a possible role in MntC function.
- Schulich Faculty of Chemistry, Technion-Israel Institute of Technology, Technion City, Haifa 32000, Israel.
Organizational Affiliation: