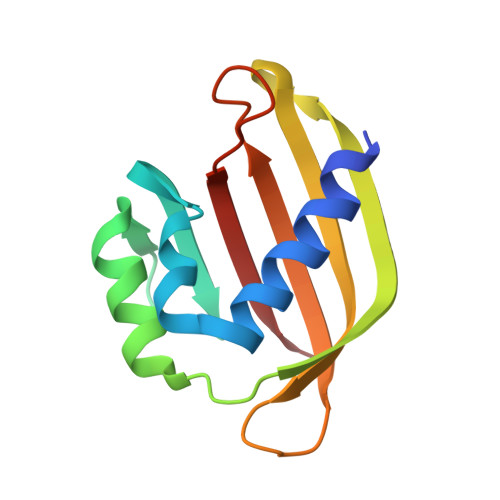
Crystal structure of the Rasputin NTF2-like domain from Drosophila melanogaster.
Vognsen, T., Kristensen, O.(2012) Biochem Biophys Res Commun 420: 188-192
- PubMed: 22414690
- DOI: https://doi.org/10.1016/j.bbrc.2012.02.140
- Primary Citation of Related Structures:
3UJM - PubMed Abstract:
The crystal structure of the NTF2-like domain of the Drosophila homolog of Ras GTPase SH3 Binding Protein (G3BP), Rasputin, was determined at 2.7Å resolution. The overall structure is highly similar to nuclear transport factor 2: It is a homodimer comprised of a β-sheet and three α-helices forming a cone-like shape. However, known binding sites for RanGDP and FxFG containing peptides show electrostatic and steric differences compared to nuclear transport factor 2. A HEPES molecule bound in the structure suggests a new, and possibly physiologically relevant, ligand binding site.
- Biostructural Research, Department of Drug Design and Pharmacology, Faculty of Health Sciences, University of Copenhagen, Universitetsparken 2, DK-2100 Copenhagen, Denmark. tv@farma.ku.dk
Organizational Affiliation: